AVIAN INFLUENZA (H9N2) VIRUSES
Avian influenza (AI) is a potentially zoonotic infection that affects poultry and may transmit to humans causing mostly self-limiting respiratory infections but can also result in multiple organ failures leading to death particularly in immunocompromised patients. Avian influenza viruses (AIV) belong to the family Orthomyxoviridae containing a negative sense single-stranded RNA genome composed of eight gene segments (PB2, PB1, PA, HA, NP, NA, M and NS). Each gene segment encodes at least one protein [Reference Yoon, Webby and Webster1]. The hemagglutinin (HA) mediates virus attachment to cellular sialic acid receptors and fusion with the host-cell membrane. A shift in receptor binding affinity from avian α2-3 to mammalian α2-6 sialic acid receptors and efficient replication in mammals of AIV can be modulated by few mutations in the HA in combination with mutations particularly in the polymerase subunits (PB2, PB1 and/or PA) [Reference Taft2–Reference Air4]. In addition, HA plays an important role in immunogenicity and, thus, successful vaccination and protection of poultry against clinical disease [Reference Cheung and Poon5, Reference Swayne6]. Influenza viruses have a high mutation rate due to the error-prone activity of the viral polymerase and therefore they change constantly. Another mechanism for virus evolution is reassortment, i.e. the exchange of gene segments between different viruses infecting the same host cell, which may lead to novel gene constellations as a prerequisite for the development of novel pandemic viruses [Reference Cheung and Poon5]. All AIV subtypes are maintained in wild aquatic birds and bird migration is the primary route for the long-range dissemination of AIV followed by introduction into domestic poultry holdings [Reference Cheung and Poon5, Reference Fouchier and Munster7, Reference Alexander8].
AIV H9N2 was first isolated from wild birds and turkeys in the USA in 1966 [Reference Homme and Easterday9]. It is the most widespread AIV subtype in poultry worldwide. Within the last two decades, H9N2 viruses were detected in wild and domestic birds, pigs, horses, minks, ferrets and humans [Reference Choi10–Reference Butt15]. In poultry, it usually causes mild clinical signs (e.g. respiratory disorders, reduced egg production and a decrease in body weight). Fatal infections occur mostly due to co-infection with bacteria and other viruses [Reference Alexander8]. The virus also induces transient immunosuppression, which may exacerbate other concomitant or secondary infections [Reference Perdue and Swayne16]. In humans, AIV H9N2 mostly causes mild respiratory illness, but fatal outcomes are sometimes observed [Reference Heidari17–Reference Uyeki21]. Although direct transmission of AIV H9N2 from birds to humans has been reported only rarely, serosurveillance studies showed that the prevalence of H9N2 infection in humans is higher than the number of confirmed cases [Reference Heidari17–Reference Uyeki21]. It is remarkable that all fatal AIVs resulting fatal infections in humans (e.g. H5N1, H7N9 and H10N8) recorded in the last two decades had acquired gene segments from H9N2 viruses [Reference Sims22–Reference Pu24]. Evolution of the virus in the last decades in poultry resulted in diversification into several genotypes. Some of them disappeared but others are still evolving [Reference Fusaro25]. Based on HA sequences, H9N2 viruses from Europe, Asia and Africa were grouped into several distinct genotypes represented by their prototype strains: A/quail/Hong Kong/G1/97 (G1-like), A/duck/Hong Kong/Y280/9 (Y280-like), A/Chicken/Beijing/1/94 (BJ94-like) and A/chicken/Korea/38349-P96323/96 (Korean-like) [Reference Guan26]. Phylogenetic analysis of all gene segments from H9N2 viruses in Asia and Europe from 1998 to 2010 revealed several genetic patterns designated as A, B, C, D with reassortment between these genotypes [Reference Fusaro25].
Little is known about the epidemiology and genetics of AIV H9N2 in the Middle East (ME) and North Africa (NA). This review, therefore, aimed to summarise the current situation of AIV H9N2 infection, evolution and control strategies in this region.
POULTRY PRODUCTION IN THE ME AND NA
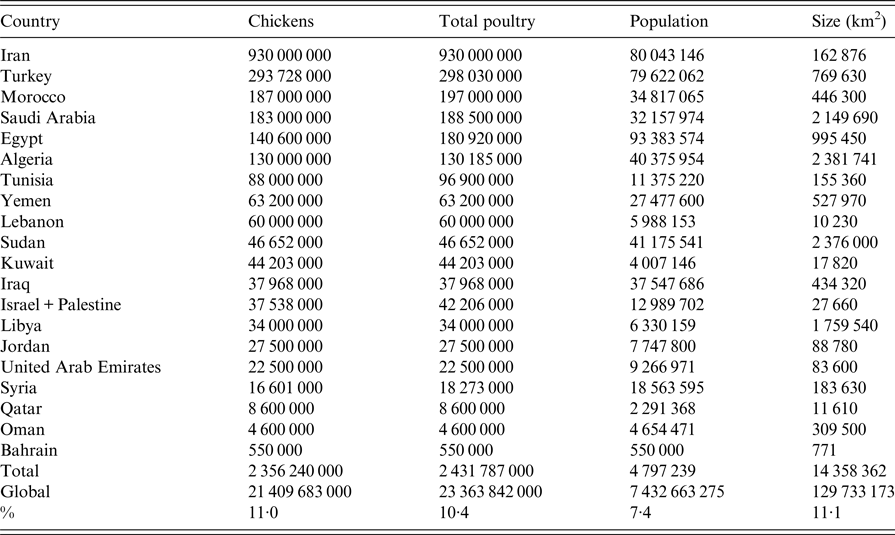
Countries in ME and NA covered in this review include Iran, Iraq, Kuwait, Qatar, United Arab Emirates (UAE), Oman, Bahrain, Yemen, Saudi Arabia (SA), Jordan, Palestine, Israel, Syria, Lebanon, Turkey, Egypt, Sudan, Libya, Tunisia, Algeria and Morocco (Fig. 1). The total human population of ME and NA was estimated in 2016 by the UN to be 0·55 billion representing 7·4% of the world population [Reference Nations27]. Egypt is the most populated country with 93 million inhabitants followed by Iran and Turkey with about 80 million each [Reference Nations27]. The ME and NA countries extend on about 14·4 million km2, representing 11·1% of the earth's surface (Table 1) and are about 3·4 times larger than the European Union [Reference Bank28]. The region produced 2·4 billion birds in 2014 representing 10·4% of the total poultry production worldwide (Table 1) according to the last reports of the Food and Agricultural Organization (FAO) of the United Nations (UN) [29]. The average annual per capita total meat consumption has more than doubled from around 12 kg in the 1990s to about 24 kg in 2010 according to USDA reports [Reference Nigatu and Motamed30]. The total growth of poultry production was estimated to be nearly 5% annually since 2000 [Reference Nigatu and Motamed31]. Iran is the country with the highest chicken production in the ME and NA and ranked number 5 in the world after China, USA, Indonesia and Brazil [29]. Likewise, Morocco and Tunisia represent number 8 and 10 for turkey production worldwide [29]. Egypt is number 10 in duck production and number 3 for geese and guinea fowl [29]. In addition, Egypt, Saudi Arabia and Jordan represent number 1, 3 and 9, respectively, for the production of pigeons and other minor birds (e.g. ostrich, quails) [29]. The region is an important market for many countries including the USA, China, Brazil, Europe, through import/export of poultry products or feed [Reference Nigatu and Motamed30]. Many pharmaceutical companies have representatives in this region. Data about the trade of poultry between ME and NA countries are scarce. The ME and NA regions are projected to import a quarter of the world's traded poultry over the period 2015–2024 [Reference Nigatu and Motamed30]. Egypt, for instance, has a self-sufficiency of table eggs but recently started to import poultry meat to cover consumption growing at almost twice the rate of production [Reference Nigatu and Motamed31].

Fig. 1. Migratory flyways of wild birds in the Middle East and North Africa.
Table 1. Poultry and human population in the Middle East and North Africa
Countries in ME and NA are located along several wild birds’ migratory flyways. Four major flyways are the Central Asia, East-Africa West-Asia, Black-Sea Mediterranean and East Atlantic routes which intersect with other pathways (e.g. East Asia-Australian flyway) (Fig. 1). Thus, millions of different species of birds fly over these countries. Stopovers of migratory birds are located in wetlands in Egypt, Jordan/Israel, Saudi Arabia, Oman, UAE, Iraq and Iran [Reference Somveille32, Reference Shobrak33]. Moreover, feral birds like pigeons, doves, ibis, sparrows and crows are frequent and widespread in these countries, which may come in contact with wild and domestic birds. In the ME, particularly in the Gulf area, raptors like falcons and Houbara buzzard are important game birds, which are also threatened by intensive hunting [Reference Somveille32]. Also, short-distance migration of some wild birds (e.g. shelduck, garganey) between countries in this region was described [Reference Gaidet34].
HISTORY AND EPIDEMIOLOGY OF AIV H9N2 IN POULTRY IN THE ME AND NA
Iran
H9N2 was detected in Iran for the first time in chickens in 1998. The outbreak caused high morbidity and mortality in broilers most likely due to mixed infection with infectious bronchitis virus (IBV), Escherichia coli, Ornithobacterium rhinotracheale and Mycoplasma gallisepticum (MG) resulting in great economic losses [Reference Karimi-Madab35–Reference Nili and Asasi37]. The clinical signs included sinusitis, facial edema, nasal and ocular discharge and severe respiratory disorders [Reference Karimi-Madab35–Reference Toroghi and Momayez38]. In 1998–2002, the virus circulated in several farms with mortality rates up to 65% [Reference Nili and Asasi36, Reference Nili and Asasi37, Reference Homayounimehr39]. Under experimental conditions the virus was shown to replicate in quails, chukar partridges and chickens with mild clinical signs and no mortality [Reference Nili40–Reference Ebrahimi43]. Efficient virus replication in dogs and mice was also described [Reference Amirsalehy, Nili and Mohammadi44, Reference Ebrahimi45]. The virus is now endemic and many reports described virus isolation or seroprevalence in chickens, turkeys and wild birds [Reference Homayounimehr39, Reference Soltanialvar, Goodarzi and Akbarnejad46–Reference Saadat53]. Vaccination was used in the field to mitigate the socioeconomic impact of the virus. However, antigenic and genetic variants evading the immune response induced by the vaccine were isolated [Reference Fereidouni51, Reference Bahari54]. In humans, up to 17% of poultry workers in different regions in Iran possessed anti-H9N2 antibodies [Reference Heidari17, Reference Alizadeh55, Reference Anvar56] and the virus replicated efficiently in human cells [Reference Shahsavandi57, Reference Farzin, Toroghi and Haghparast58]. Iran is located on two major flyways, the central Asian and Black Sea-Mediterranean flyways [Reference Gaidet34], which may be important for the introduction of AIV, including H9N2, into the ME and NA.
Iraq
Since 2004, H9N2 is endemic in poultry in Iraq vaccination is used intensively to control the disease [Reference Kraidi59, Reference Kraidi60]. In 2004–2007, Iraq experienced many H9N2 outbreaks with mortality rates up to 70% in broilers and 10% in layers and breeders [Reference Khamas61]. From June to December 2008, the virus spread widely in poultry in Iraq where 100% (53/53) of broiler flocks in Najaf province with history of respiratory signs were positive for H9N2 RNA [Reference Al-Mohana62]. From November 2010 to June 2011, 47·3% (18/38) of broiler chickens in many regions of Iraq were positive by polymerase chain reaction (PCR) [Reference Al-Dabhawe, Kadhim and Samaka63]. AIV H9N2 infection was mostly accompanied by Newcastle Disease Virus (NDV) infections [Reference Al-Mohana62]. From July 2012 to July 2013, 60 out of 251 flocks were positive by RT-qPCR in Al-Najaf-Iraq [Reference Al-Kelaby, Kadhim and Ghazzay64]. From September 2013 to June 2014, H9N2 was detected in broiler chickens suffering from respiratory signs and in asymptomatic wild birds from southern provinces of Iraq [Reference Abdul-Sada65]. From September 2014 to June 2015, H9N2 RNA was detected in 16 out of 100 broiler flocks showing respiratory signs from seven provinces in the middle and southern regions of Iraq. Wild birds were blamed for the introduction of the virus into Iraq [Reference Kraidi60]. Also, in the same period in 2014–2015 the authors described the isolation of six H9N2 viruses from broiler flocks in five provinces situated in the Middle and Southern parts of Iraq [Reference Kraidi59].
Kuwait
The virus was isolated from chickens in 2003–2005 [Reference Alexander66, Reference Slomka67] and in 2008 [Reference Brown68]. The vaccine is also known to be marketed in Kuwait [69].
Qatar
In 2008, an H9N2 was isolated from poultry in Qatar [Reference Fusaro25]. However, no data on virus epidemiology are available.
United Arab Emirates
The first isolations of AIV H9N2 in UAE were reported in 1999 from chickens and from other birds in 2000 [Reference Alexander66, Reference Alexander70, Reference Banks71]. Between 2000 and 2003, H9N2 viruses were isolated from farmed quails and chickens. Viruses from quails caused a 30% decrease in egg production without producing overt clinical signs, whereas the chicken isolates caused rapid mortality (up to 36%) due to respiratory disorders [Reference Aamir72]. After experimental infection, chickens did not show clinical signs although viruses were successfully transmitted to contact animals. In mice, after intranasal inoculation few signs of weight loss and morbidity were observed but the virus was not detected in the brain, spleen, or blood of infected mice. Moreover, all isolates showed high resistance to amantadine [Reference Aamir72]. In 2005–2011, viruses were isolated from diseased hosts such as chickens, pheasant, stone curlew, White-Bellied Buzzards and quails [Reference Wernery73]. Under experimental conditions, chickens inoculated with several of these viruses showed no clinical signs but the viruses replicated to higher titres in the respiratory tract than in the intestinal tract [Reference Wernery73]. In 2006–2007, the virus was isolated from dove, Houbara, quails and falcon [74]. In 2015, the virus was isolated from vaccinated 60-week-old chickens [Reference Lau75].
Oman
Few research publications about AIV H9N2 infections in Oman exist. In 2005–2006, 18 (9·4%) out of 192 swab samples collected from chickens and doves were classified as AIV H9N2 [76]. Also, AIV H9N2 was isolated from free-living birds (mynah, Acridotheres tristis) of the starling family and from chickens. Both viruses were closely related genetically assuming introduction of the virus into poultry by wild birds [Reference Body77]. The virus exhibited low pathogenicity in chickens with an IVPI of 0·04 and four experimentally inoculated birds showed only mild respiratory disease and ruffled feather on days 6 and 7 post-inoculation [Reference Body77]. In 2012, serum and swabs collected from 2350 birds of 243 backyard flocks (including chickens, turkeys, guinea fowl, ducks and geese) from all regions and governorates of Oman indicated a wide distribution of AIV. About 38% of tested flocks were seropositive by ELISA but no virus was detected by RT-PCR [Reference Shekaili78]. Vaccination of commercial poultry against AIV H9N2 is implemented in Oman [69, Reference Shekaili78].
Yemen
H9N2 is enzootic in poultry in Yemen and vaccines are in use [69, 79]. However, no data are available on virus epidemiology and genetic traits.
Kingdom of Saudi Arabia (KSA)
KSA was the first country to isolate AIV H9N2 in the region where infection was reported in chickens in 1998 [Reference Alexander70]. The virus was isolated from chickens in 1999 [Reference Banks71], 2002 (Kim et al. unpublished), 2003–2005 [Reference Alexander66] and in 2005, 2006 and 2010 [Reference Slomka67]. In 2006–2007, serosurveillance and molecular detection in broilers (n = 1561), layers (n = 988), ducks (n = 329) and pigeons (n = 450) revealed widespread AIV H9N2 infection particularly in the Northern regions [Reference Alkhalaf80]. Vaccination of poultry in KSA is widely used and infections in vaccinated birds are not uncommon [Reference Gharaibeh and Amareen81, Reference Bakri82].
Jordan
Serosurveillance using indirect ELISA in 38 clinically healthy breeder-broiler farms performed from October to December 2001 in Southern Jordan revealed 71% positive flocks. They were mostly located within the migratory flyway corridor [Reference Al-Natour and Abo-Shehada83]. In 2003–2005, the virus was isolated from chickens and domestic ducks [Reference Alexander66, Reference Slomka67, Reference Monne84, Reference Gharaibeh85]. An infected broiler flock in 2003 suffered 30% mortality. Under experimental conditions, inoculated broilers showed respiratory signs and loss in body weight and excreted the virus for up to 9 days although none of the chickens died [Reference Gharaibeh85]. In ferrets, the virus replicated efficiently in inoculated animals but was not transmitted to their contacts [Reference Wan86]. In 2006–2007, 46 outbreaks in chickens were recorded [Reference Brown68]. From 2005 to July 2007, 115 non-vaccinated commercial broiler chicken flocks that suffered from respiratory disease were investigated. Antibodies against AIV were detected in 15·7% of the surveyed flocks and usually accompanied by NDV or IBV [Reference Roussan, Haddad and Khawaldeh87]. February 2006 to November 2007, Roussan et al. [Reference Roussan88] found anti-AIV H9 antibodies in 65 out of 120 broiler flocks (54·2%) and 47 out of 60 layer flocks (78·3%). Viral RNA was detected in 31 out of 60 broiler flocks (51·7%) and 15 out of 23 layer flocks (65·2%). In 2011–2015, the virus was reported in 83 out of 350 (23·7%) non-vaccinated poultry flocks using RT-PCR [Reference Roussan, Khawaldeh and Shaheen89]. The infection was very common in the broiler flocks and also concomitant was MG or Mycoplasma synoviae (MS) [Reference Roussan, Khawaldeh and Shaheen89]. In 2013, a genetic drift H9N2 virus deviating from the vaccine strain was isolated from vaccinated broilers flocks with a history of increased mortality and severe clinical signs [Reference Gharaibeh and Amareen81]. The HA gene of the isolated virus was only 89·1% identical to the vaccine strain HA gene. Serum antibodies elicited by the classical vaccine from 2004 had low cross-reactivity against this virus indicating significant antigenic drift. Therefore, update of the H9N2 vaccines in Jordan was recommended to increase protection levels [Reference Gharaibeh and Amareen81].
Israel
In Israel, there were several introductions of AIV H9N2 into poultry via wild birds (Panshin et al. unpublished) or from neighboring countries (Shkoda et al. unpublished). The first isolation of H9N2 from chickens and turkeys in the Northern part of Israel occurred in 2000 [Reference Alexander66, Reference Perk90]. In 2001, outbreaks of H9N2 were detected in two turkey flocks in central regions and in 2002–2003 the infection spread to the Northern and Southern regions infecting mainly turkeys and chickens. Affected birds showed mild to severe respiratory signs, edema of the head and face and decreased egg production with varying mortality between 0 and 30% according to the type of secondary bacterial infection [Reference Perk91]. From 2000 to 2006, over 500 H9N2 viruses were isolated from different poultry species [Reference Banet-Noach92–Reference Perk94]. In 2006–2007, the virus was reported in chickens and turkeys [Reference Brown68]. The most recent introduction of H9N2 into poultry in Israel was in 2016 from Egypt (Shkoda et al. unpublished). Since 2000, the virus infected chickens, turkeys, geese, ostriches and wild pigeons [Reference Perk91, Reference Banet-Noach92, Reference Davidson95]. Infection of pigs in 2009–2011 was not detected [Reference Davidson96]. Under experimental conditions, some H9N2 viruses isolated from Israeli turkeys were avirulent in chickens [Reference Khalenkov97]. The severe damage to poultry was most likely due to co-infection, which is not uncommon in the field [Reference Perk91]. To control the disease, at least two vaccines were used in Israel. The first vaccine contained A/turkey/Israel/965/02 and has been used since 2003, while the second vaccine containing A/Chicken/Israel/215/07 was introduced at the end of 2008 [Reference Davidson95, Reference Banet-Noach98]. Vaccination was claimed to accelerate the evolution of the H9N2 viruses in Israel [Reference Davidson95].
Lebanon
In 2004–2005, AIV H9N2 was isolated from chickens in different sectors [Reference Alexander66]. The mortality rate was up to 35% in broilers and 1–2% in breeders and layers [Reference Barbour99]. The virus was detected in the brain of broilers and up to 72% drop in egg production was reported [Reference Barbour99]. Interestingly, pigs fed on dead H9-infected chickens developed anti-AIV antibodies. Also, one third of poultry farmers in this survey seroconverted without showing clinical signs [Reference Barbour99]. The virus showed low pathogenicity in hamsters [Reference Shaib100], while in chickens the virulence increased after several passages in embryonated eggs or chickens [Reference Shaib101, Reference Shaib102]. An H9N2 virus in Lebanon was resistant to oseltamivir [Reference Murtada, Barbour and Shaib103]. In 2010, two viruses were isolated from quails (Webby et al., unpublished). Vaccination against H9N2 is also applied in Lebanon [69].
Turkey, Syria and Bahrain
There are no reports on the prevalence of AIV H9N2 in Turkey, Syria and Bahrain, although vaccines are sold there [69].
Egypt
The first report of AIV H9N2 in Egypt was in 2006 from live poultry markets using real-time RT-PCR. However, no virus isolation was reported (Spackman et al., unpublished data). From February 2009 to April 2012, antibodies against H9 viruses were widespread in poultry in Egypt [Reference Afifi104]. The earliest virus isolation of the current H9N2 outbreaks in Egypt dates from December 2010. The virus was strikingly different from the first introduction in 2006. Since then, AIV H9N2 infected a wide range of birds in Egypt including chickens, quails, ducks, turkeys and pigeons in commercial and backyard sectors [Reference Kandeil105–Reference Awad, Arafa and Hagag111]. Most of infected chickens and turkeys exhibited respiratory distress and/or decrease in egg production, but some quails and broilers flocks showed no overt symptoms [Reference Afifi104–Reference Arafa106, Reference Monne108, Reference El-Zoghby110]. The majority of outbreaks are reported during the winter months but outbreaks are observed year-round particularly in the Nile Delta [Reference Arafa106, Reference Abdelwhab and Abdel-Moneim112]. Co-infections of poultry with other viruses (e.g. IBV, NDV, H5) or bacteria (MG, MS) are common. Interestingly, out of 86 broiler flocks 42% were co-infected with H9N2 and IBV and in ⩽1% a mixed triple infections with IBV-H5-H9-NDV was observed in 2012–2014 [Reference Hassan113, Reference Naguib114]. To date, no reassortment between the co-circulating H5N1 and H9N2 viruses has been reported. Inactivated vaccines using local and non-local field strains of H9N2 are frequently used in Egypt and the emergence of antigenic drift variants has been reported. It is worth mentioning that the Egyptian viruses reacted poorly against serum samples from the ME including vaccination derived sera [Reference Kandeil105, Reference Adel115, Reference Kandeil116] (Naguib et al. unpublished). In pigs, H9N2 infection was widespread as observed by serological investigation in 2014–2015 (Gomaa et al. unpublished). In 2015, three children with a history of exposure to poultry were found positive for AIV H9N2 RNA [Reference Abdelwhab and Abdel-Moneim112] and up to 7·5% seroprevalence in exposed humans was reported [Reference Gomaa117]. Human infections showed transient influenza like illness but subsided without sequelae.
Sudan
The authors failed to find any reports on the prevalence of H9N2 in Sudan.
Libya
So far, there are two independent incidences of H9N2 viruses in Libya. The first event was the detection of the virus in commercial poultry in 2005–2006 [Reference Alexander66, Reference Slomka67]. The second occurred in 2013, when the virus was successfully isolated from layers, broilers chicken flocks and peacock [Reference Kammon118]. The infection spread rapidly all over the country and the flocks were simultaneously co-infected with NDV. Birds in the affected flocks showed respiratory signs and high mortality [Reference Kammon118]. There are no reports of application of H9N2 vaccines in the field, but vaccines are marketed in Libya [69].
Tunisia
H9N2 viruses have emerged in Tunisia in 2009 causing several outbreaks in poultry flocks [Reference Tombari119]. AIV H9N2 virus was also isolated from wild birds [Reference Tombari120]. Moreover, a nationwide serosurvey of 800 flocks in 2010–2011 indicated widespread AIV infection including H9N2. A total of 223 flocks had anti-NP antibodies (28·7%) particularly in the coastal areas during the autumn and winter. The infection was higher in layer and breeders flocks than broilers. A total of 20 isolates were confirmed by RT-qPCR. Low biosecurity measures and contact to wild birds were claimed to be the source of infection [Reference Tombari121]. In 2012, H9N2 was isolated from a broiler flock [Reference Aouini, Laamiri and Ghram122]. Infection of poultry has also been recorded in 2014 (Arbi, unpublished data). Antivirals zanamivir and amantadine decreased virus replication in experimentally inoculated chickens [Reference Umar123].
Algeria
To date there are no data available on the prevalence of H9N2 in Algerian poultry [Reference Sid, Benachour and Rautenschlein124]. No H9 antibodies were detected in samples obtained from broilers, turkeys or layers flocks in February 2012 and August 2013. It was mentioned that no vaccination against AIV is implemented in Algeria [Reference Sid, Benachour and Rautenschlein124].
Morocco
The first outbreak of H9N2 in Morocco was reported in January 2016 in broilers and breeders [Reference El Houadfi125]. Within few weeks, the virus spread to several locations in the country infecting chickens in layers and breeders farms inducing decrease in feed consumption, severe respiratory signs and mortality rates from 2 to 15%. The outbreaks were associated with a sharp drop in egg production (up to 80%) with no complete recovery after 10 weeks of infection. Similar clinical signs were reported in turkey flocks with mortality rates around 10% [Reference El Houadfi125]. Emergency vaccinations of poultry in all production sectors were implemented. The officials announced the control of the virus by April 2016 [Reference El Houadfi125]. Possible routes for introduction of the virus were inconclusive.
GENETIC RELATEDNESS OF H9N2 IN THE ME AND NA
To assess the relatedness of H9N2 viruses in the ME and NA, a phylogenetic tree based on HA gene sequences (n = 760) retrieved from GenBank and GISAID in March 2017 (Fig. 2, Supplementary Fig. S1 and Table S1) was calculated. Genetic analysis of all HA gene sequences from the ME and NA indicated diversification of the H9N2 viruses in this region. Two major lineages were observed. Lineage A represents recent viruses from 1998 to 2016 in all countries of the ME and NA, whereas lineage B represents early viruses from 1998 to 2007 in Saudi Arabia, Iran and Israel. Lineage A most likely originated from Pakistan in 1998. It contains major genetic groups including the Egyptian viruses, Israeli viruses, panzootic viruses designated Middle East 1 (ME1) and Pakistani-Iranian viruses. The Egyptian (EG) viruses were introduced in 2010 probably from Israel. Continuous evolution of the EG viruses mostly under vaccination pressure resulted in rapid diversification of three groups (EG1 to EG3), where two groups (EG1 and EG2) co-circulated together from 2010–2011 to 2015. EG3 contains viruses isolated from 2015–2016 in Egypt which spilled over to poultry in Israel in 2016. The Israeli viruses are divided into three major distinct groups (designated IS1 to IS3) mostly following a temporal pattern from 2007 to 2013 as previously found [Reference Davidson95]. Viruses in group IS1 were reported in 2010 in poultry in Lebanon and Jordan. Another group of viruses in lineage A is a panzootic group ME1. This panzootic group was reported in KSA in 2005 to 2010, Libya in 2006–2013, Qatar in 2008, UAE in 2008–2015, Tunisia in 2010–2014, Israel 2011–2016 and Morocco 2016. Moreover, two Pakistani-Iranian groups (PA-IR1 and PA-IR2) were clustered separately. PA-IR1 contains viruses isolated from poultry in Kuwait in 2004, Pakistan in 2008–2012, Iran in 2010–2016 and Iraq in 2014, while PA-IR2 contains viruses isolated from Pakistan in 2004–2008 and descendent viruses from Iran 2009 to 2014. Furthermore, viruses isolated from UAE in 1999–2002 and Oman in 2005–2006 clustered in Gulf1 group, whereas viruses isolated from UAE, Iraq, Iran and Pakistan in the period from 2003 to 2015 allocated in Gulf2 group.

Fig. 2. Phylogenetic relatedness of the HA gene sequences of H9N2 viruses in the Middle East from 1998 to 2016. HA gene sequences were collected from countries in the Middle East in addition to Pakistan. A total of 760 sequences were retrieved from the GenBank and GISAID and aligned using MAFFT and were further edited using Bio Edit. The phylogenetic tree was generated by IQTREE using the best fit model selection. The tree was further edited for publishing using FigTree and Inkscape. Two major lineages were observed, lineage A and lineage B. Lineage A contains viruses from Egypt (EG1, EG2 and EG3; red) in addition to viruses from Israel (IS1, IS2 and IS3; green), the Middle East group with viruses from Israel (green), KSA (magenta), UAE (cyan), Tunisia, Morocco and Libya (black), the Pakistani-Iranian groups (PA-IR1 and PA-IR2) with viruses from Iran (blue) and Pakistan (yellow) and, finally Gulf1 and Gulf2 representing viruses from UAE, Oman, Iraq, Iran and Pakistan. Lineage B contains three genetic groups IR1, IS4 and IS5. All viruses in lineages A and B belonged to the G1-like H9N2 lineage, except IR2 which was similar to the Korean-like lineage.
Lineage B is mostly limited to the Asian part of the ME and contains three distinct clusters IR1, IS5 and IS6. IR1 contains viruses from Iran in 1998 to 2007. The root of this group is a virus from chickens in KSA in 1998. IS4 contains the earliest viruses from Israel in 2000–2003, which share an ancestor with viruses from KSA in 1998. Interestingly, this virus probably transmitted back to quail in KSA in 2006, or the ancestor virus circulated unnoticed in parallel in KSA. IR2 represents a separate introduction from wild birds in 2007 by a virus closely related to viruses from China (data not shown). IS5 contains viruses from Israel in 2003–2007, which were also isolated from different birds in Jordan in 2004 and 2007 and Lebanon in 2004. All viruses in the ME and NA belonged to the G1-like lineage except viruses from wild birds in Iran in 2007 (IR2 group) [Reference Fereidouni51], which were closely related to the Korean-like lineage.
To study possible reassortment within different lineages in the ME and NA, 95 full genome sequences retrieved from the GenBank were analysed by MrBayes. Interestingly, several reassortment events have been observed (Supplementary Fig. S2). Two Egyptian viruses from pigeons in 2014 (A/pigeon/Egypt/S10408B/2014 and A/pigeon/Egypt/S10409A/2014) acquired PB2, PB1, PA, NP and NS from viruses closely related to those circulating in Iran, Pakistan, Lebanon and Gulf area [Reference Kandeil116]. Likewise, two viruses from quails in Lebanon in 2010 (A/Quail/Lebanon/272/2010 and A/Quail/Lebanon/273/2010) acquired their NP, M, PB2 and PA genes from the same area as the two Egyptian viruses. Finally, an Iranian virus (A/Chicken/Iran/ZMT-101/1998) is most likely a reassortant as well (Supplementary Fig. S2). Many mutations in AIV H9N2 in ME and NA associated with adaptation to mammals have been described [Reference Fusaro25, Reference Guan26, Reference Soltanialvar, Goodarzi and Akbarnejad46, Reference Bashashati, Vasfi Marandi and Sabouri48, Reference Moosakhani50, Reference Kraidi59, Reference Lau75, Reference Monne84, Reference Davidson95, Reference Kandeil105, Reference Kammon118, Reference Tombari119].
CONCLUSION
AIV H9N2 is enzootic in poultry in the ME and NA. While reports are adequate for Egypt, Iran, Israel and Jordan, little is known about the epidemiology of the disease and genetic features of H9N2 in the other countries. Ongoing infection was reported despite intensive vaccination of poultry [Reference Gharaibeh and Amareen81]. Thus, it appears essential to routinely update the vaccine strains. Regional surveillance may be useful to select the best vaccine candidates. AIV H9N2 infection was mostly accompanied and exacerbated by co-infecting MG, MS, IBV, NDV, AIV H5N1 or bacteria. AIV H9N2 infection was also reported from pigs in Egypt and Lebanon and humans in Egypt, Iran and Lebanon. Genetic analysis showed the dynamics of the H9N2 viruses in ME and NA. Some lineages remained localised within the country and rarely crossed national borders, while others spread to other countries with a panzootic group isolated from many countries in the Gulf area, Israel and North Africa. A vaccine strain from this group may be useful to control the virus in these countries. The current Israeli 2015–2016 viruses are mixtures of several lineages which may be a problem for vaccine production. Also, in Iran in addition to the extinct IR1 and the exotic IR2, there are three different groups circulating for a period of time, PA-IR1, PA-IR2 and Gulf2. Reassortant viruses were also observed probably due to movement of infected domestic poultry from area to another or contact to wild birds. Finally, the H9N2 in the ME and NA continue to cause losses in vaccinated and non-vaccinated poultry, carry genetic features of mammal-adaptation [Reference Wan86], have been already isolated from humans and co-circulate with other AIV (e.g. H5N1 and H5N8). Thus, the enzootic H9N2 in the ME and NA merit in-depth investigation including regional surveillance and control activities, revision of used vaccines, surveillance in human populations and other animals. A notification platform or website for the disease and infection in the ME and NA may be useful. Unravelling the pathobiology and genetic features of the circulating H9N2 viruses is important to assess the risk for public health.
SUPPLEMENTARY MATERIAL
The supplementary material for this article can be found at https://doi.org/10.1017/S0950268817002576






