Introduction
Environmental exposures during prenatal life, such as maternal diet, alcohol/drug abuse, smoking, preeclampsia, gestational diabetes, and psychosocial stress, may result in sustained effects in the molecular mechanisms programing offspring health, and contribute to neurodevelopmental disorders risk. Reference DeSocio1 Epigenetic mechanisms are associated with environmental stimuli, especially DNA methylation (DNAm), which has been suggested as a potential biomarker of and mediator in health outcomes. Reference Felix and Cecil2-Reference Lee, Kalia and Frederica Perera4 It was previously reported that the environmental input can be mediated by DNAm on important genes associated with neural circuitry components, thereby modulating brain plasticity, Reference Miller5 but evidence is still lacking as to whether the relationship between neurodevelopmental outcomes and prenatal interventions are mediated by modifications in DNAm.
Observational studies in humans have found that specific patterns of DNAm are associated with stressful prenatal environments and specific behavioral outcomes of the child. Reference Neumann, Walton and Alemany6,Reference Han, Patel and Jones7 For example, differential methylation of specific sites of the BDNF gene in cord blood, placental tissue, and maternal venous blood were associated with exposure to war trauma and chronic stress during gestation. Reference Kertes, Bhatt and Kamin8 Indeed, prenatal famine exposure predicted changes in metabolic markers in adulthood and were mediated by DNAm in genes related to the metabolism. Reference Tobi, Slieker and Luijk9 DNAm levels of specific single CpG sites (CpGs) and/or entire regions are frequently associated with prenatal stress and brain development, indicating the relevance of exploring this mechanism. Reference Rijlaarsdam, van IJzendoorn and Verhulst10,Reference Viuff, Sharp and Rai11 At the same time, a recent development is that the estimation of gestational age using DNAm levels of specific CpGs has been validated and has been associated with prenatal stress. Reference Girchenko, Lahti and Czamara12 However, these biological outcomes have not been studied in longitudinal studies using relevant infant health outcomes.
Considering the concept of tissue plasticity during intrauterine life, psychosocial interventions during pregnancy have been found to improve conditions for the child’s development. Reference Britto, Lye and Proulx13 Indeed, a recent meta-analysis reported that psychosocial interventions in early pregnancy promote positive developmental outcomes. Reference Aboud and Yousafzai14 Nevertheless, there are only a few randomized controlled trials testing psychosocial interventions during pregnancy designed to determine the effect of maternal stressors on the epigenetic mechanisms that promote fetal neural development. Reference Fisher, Beauchamp, Roos, Noll, Flannery and Delker15 One key example is the Nurse Family Partnership program that found variations in DNAm in the blood of offspring at 27 years of age, independent of sex, ancestry, cellular heterogeneity and a polygenic risk index for major psychiatric disorders, but dependent of some lifestyle factors, such as smoking. Reference OʼDonnell, Chen and MacIsaac16 Still, there are a few conceptual nuances that are not explained in these studies that could be investigated in a randomized trial of pregnant women.
Indeed, there are relatively few studies that have integrated clinicals with epigenetic and phenotypic data to explicitly explore the role of DNAm as a potential biological mediator of environmental effects on health outcomes. Reference Felix and Cecil2,Reference Barker, Walton and Cecil17 While most of the research on this topic was conducted in high-income countries, the effects may be different and possibly even greater in low- and middle-income countries given the high degree of stress due to living in conditions with high levels of interpersonal violence, natural disasters, poverty, and exposure to infectious diseases, Reference Goldstein, Norris and Aronoff18 food insecurity, dietary deficiencies, crowded housing, and extremes in temperature. Reference Herba, Glover, Ramchandani and Rondon19 To address this important gap in the literature, we conducted a study to investigate whether “Primeiros Laços,” a program that provided support to pregnant teenagers living in extreme social vulnerability would promote epigenetic changes at CpGs, regions and network modules as well as differences in age acceleration (AA), represented here by the difference of methylation age and birth age). Then we performed a potential causal mediation analysis using methylation results as potential mediators for neurodevelopmental outcomes of offspring at 12 months of age.
Method
Study design
A randomized controlled trial was conducted to test the effects of the “Primeiros Laços” program in improving mother-infant relationship (NCT02807818). Reference Fracolli, de Reticena, de Abreu and Chiesa20
Study population
The study included first-time pregnant adolescents living in an urban area characterized by high rates of poverty and urban violence in Sao Paulo, Brazil. Eighty women were randomized to the intervention (n = 40) and to the control (received the usual prenatal care, n = 40) groups. The inclusion criteria were (1) women between 14 and 19 years that were (2) pregnant for the first time, (3) between 8 and 16 weeks’ gestation, (4) living in western regions of São Paulo, and (5) low socioeconomic status [classes C–D/E] according to the Criteria for Economic Classification assessed by the Brazilian Association of Research Companies questionnaire. 21 Exclusion criteria were (1) mental, visual, or hearing disabilities. Randomization was stratified according to type of basic health unit and years of grandmother’s schooling. In Brazil, basic care, considered the preferential path of access of the population to Unified Health System services, oversees the organization and integration of public healthcare networks.
Intervention
Primeiros Laços is a home-visit program using visits from trained nurses to first-time pregnant adolescents and their infants from the first 16 weeks of pregnancy until the child is 24 months old. The nurses specialized in maternal or mental health and were supervised weekly by senior nurses and psychologists. Primeiros Laços is based on three theoretical frameworks (Attachment theory, Bowlby et al., 1979; Self-Efficacy theory, Bandura, 1977; Bioecological Development theory, Bronfenbrenner, 1998) and was structured in five axes (health care, health environment, parenting and attachment, social and family network, life project). The program was developed by our team (Chiesa et al., 2008) based on existing home-visiting programs (Olds, 2006; Slade et al., 2005). The care plan for each mother–infant dyad was carefully designed to strengthen maternal competences for warm and responsive care. A key factor was establishing positive relationships between home visitors and the family. During the visits, nurses helped parents develop child-centered interactions, improve their bond with their infant, reflect on their own attachment history and parenting they received, consider their child as an individual with his/her own needs, feelings, and thoughts, and improve their parenting skills by modeling. The nurse encouraged attuned parenting and stimulated sensitive behaviors, such as being attentive to the child’s communicative signals or following the child´s lead. Parents were also given support to think in a reflective way. Frequency of visits was weekly (first/last month of pregnancy/puerperium), biweekly (gestation/2–20 months of child’s age) and monthly (21–24 months of child’s age). Mothers were expected to receive 60–62 visits in total.
Care-as-usual
Control participants received care-as-usual from the Sistema Único de Saúde, Brazil’s public health system, Reference Castro, Massuda and Almeida22 according to national guidelines of the Ministry of Health that are in line with the World Health Organization (WHO) guidelines. Prenatal and postnatal care is delivered by primary healthcare units free of charge and focuses on preventive interventions, early detection of gestational risk, and referral to specialized health services in case of high-risk pregnancies. Participants from the intervention group also had access to these public health services.
Behavior and neurodevelopmental outcomes
To assess socioeconomic conditions, the following instruments were used: Brazilian Scale of Food Security (EBIA), Reference Pérez-Escamilla, Segall-Corrêa, Maranha, de Fátima Archanjo Sampaio, Marín-León and Panigassi23 monthly family income, and maternal education. Data for alcohol/drug abuse and smoking habits were also collected. Anthropometric data were used for nutritional status evaluation. To evaluate depression and anxiety, the Beck Depression and the Anxiety Inventory (BAI), Reference Quintão, Delgado and Prieto24,Reference Gomes-Oliveira, Gorenstein, Neto, Andrade and Wang25 were collected at baseline and at 30th gestation week. Infant weight and length were measured at birth by trained professionals. The values (z-score) of the infant’s growth parameters were based on the growth standards of the WHO. Reference de Onis, Garza, Onyango and Rolland-Cachera26
Finally, the Bayley Scales of Infant Development (BSID-III) Reference Madaschi, Mecca, Macedo and Paula27,Reference Bayley28 were conducted by trained psychologists at 12 and 24 months of age to evaluate neurodevelopmental characteristics of the infants. The five neurodevelopmental domains were considered separately: Cognitive, Receptive Language, Expressive Language, Fine Motor Skills and Gross Motor Skills.
Sample processing to collect biological samples
The mothers were advised to contact researchers when going into labor, however some mothers forgot to notify them when the process began. In addition, others gave birth outside the referral hospital, as they were geographically far away when they went into labor. Considering these challenges, we were able to collect biological samples from 15 women from the control and 17 intervention groups.
From 32 neonates, 4 ml of umbilical cord blood were collected in EDTA vacutainer tubes (B.D. Scientific) immediately after delivery and the samples were immediately frozen at −80°C for following analyses. DNA was extracted with the QIAamp DNA Mini Kit (Qiagen). NanoDrop (Thermo Fisher Scientific) and Qubit (Thermo Fisher Scientific) were used to assess DNA purity and quantity, respectively. Bisulfite converted DNA (EZ DNA Methylation kit, Zymo Research Corp) was hybridized in the HumanMethylation450 BeadChip arrays (HM450K, Illumina), following the Illumina Infinium HD methylation protocol. Both experiments were processed by Deoxi Biotecnologia (www.deoxi.com) according to the manufacturer’s instructions. Raw data were extracted by the iScan SQ scanner (Illumina) using GenomeStudio software (v.2011.1), with the methylation module v.1.9.0 (Illumina), into IDAT files, which were used for further analyses.
Minfi package version 1.26.2 Reference Aryee, Jaffe and Corrada-Bravo29 was used for quality control of the samples as well as to apply quantile normalization Reference Fortin, Triche, Hansen and Hancock30 and to calculate M-values. We removed probes that contained SNPs at the CpGs, those located at the sex chromosomes, and those known to cohybridize to more than one genomic sequence resulting in 429,174 probes. ChAMP package version 2.10.2 Reference Tian, Morris and Webster31 was used to identify and adjust the M-values for batch effects and for sex.
Differential methylation analysis
Differential methylation positions- CpG sites (DMPs) and differentially methylated regions (DMRs) were calculated from M-values. Differentially methylated CpGs (adjusted p-value ≤ 0.05, Benjamini–Hochberg method) and DMRs (False Discovery Rate, adjusted p-value ≤ 0.05) were obtained using the DMP and DMRcate functions Reference Peters, Buckley and Statham32 respectively, as implemented in ChAMP. After identifying the DMPs, they were further annotated to obtain information regarding enhancers, transcription factor binding sites (TFBSs) and DNAse I Hypersensitivity sites (DHSs). For enhancers, we used the complete annotations from EnhancerAtlas for Fetal Brain and Fetal Placenta. For TFBSs and DHSs we used the wgEncodeRegDnaseClusteredV3 and wgEncodeRegTfbsClusteredV3 tracks from Genome Browser’s Data Integrator (available at http://genome.ucsc.edu/cgi-bin/hgIntegratorhttp://genome.ucsc.edu/cgi-bin/hgIntegrator). We considered DMRs sequences of 300 bp containing at least seven CpGs and showing differences of FDR ≤ 0.05.
Weighted correlation network analysis
The weighted correlation network analysis package Reference Langfelder and Horvath33 was used to compare comethylation networks from intervention and control groups using the previously identified DMPs. For each group, Pearson correlation between samples M-values were used to construct two correlation matrices, with the estimated network power value (beta) = 15 was used as a scale-free independence index, resulting in an R 2 of 0.90 and 0.96 for interventional and control groups, respectively. For both networks, we used the following parameters: deepsplit = 3, mergecuthigh = 0.25, and minimum module size = 30.
Z-summary was used to evaluate preservation between networks (100 permutations). Modules with a low or moderate level of preservation are those with Z-summary ≤ 10, which indicates large topological variation. Reference Langfelder, Luo, Oldham and Horvath34 Considering that the Z-summary indicates a topological variation between each group, modules with a low Z-summary value indicate the CpGs with the largest differences in the comethylation pattern. CpGs with a kME correlation ≥ 0.9 were considered hubs of that module.
Epigenetic clock for gestational age at birth
As proposed by Knight et al., Reference Knight, Craig and Theda35 an Epigenetic Clock for Gestational Age at birth was calculated by submitting the DNAm levels of 148 CpGs sites to an elastic net regression based on six training datasets.
Statistical Analyses
Statistical analyses were performed in R v. 3.5.0. Chi-square and t-tests were applied to compare the phenotypes between intervention and control groups. False Discovery Rate, Benjamini−Hochberg and bootstrapping (1000 repetitions) was used to multiple comparations (Adj p-value < 0.05).
Due to 30% of neurodevelopment outcomes missing data from our sample at 24 months of age, we only use outcomes at 12 months of age to perform the proposed analysis. Moreover, 3 of 32 cord blood samples did not have neurodevelopmental outcomes collected at 12 months, therefore we analyzed 14 and 15 from control and intervention, respectively.
Thus, although the cognitive and the gross motor skills were different between intervention and control groups in this subsample with cord blood collection, we only analyzed the cognitive score as an outcome because it was also statistically significantly between cases and controls in the whole cohort at 12 months of age. RCT Primeiros Laços had 63 neurodevelopmental evaluations at 12 months of age (Suppl. File 1. Supplementary Table 2). The study design can be visualized in Suppl. File 1 (Supplementary Fig. 1).
To perform the mediation analysis, we defined by the following criteria at CpGs, region, or module level: (1) Only DMPs associated with cognition were tested as mediators. DMPs related to cognition (R 2 adj > 0.4) were defined by a linear regression model using M-values of DMPs as dependent variable and the cognitive domain score controlled by sex. (2) From coexpression data, all CpGs that were hubs from the not preserved comethylation module. (3) Each DMR was represented as the M-values means of all CpGs within the specific region. (4) Epigenetic clock as raw values.
We used the mediation R package version 3.5.0 Reference Tingley, Yamamoto, Hirose, Keele and Imai36 to analyze causal mediation with bootstrap of 1000 replications for confidence intervals. Data from mediation model for each CpGs or DMRs include estimates and 95% CIs of total, direct, and mediation effects. The average causal mediation effects (ACME) states whether intervention positively (the probability of increasing cognition score) or negatively (decreases the cognitive score) affects the outcome (cognitive score) through a mediator (DNAm, according to the selected ones). Total effect and average direct effect (ADE) show if the intervention is likely to change cognition in the mediation models. The function medsens was applied to perform sensibility analysis to test effects of unmeasured confounders. A p-value less than 0.05 was considered statistically significant.
Results
Characteristics of the sample
Cord blood was collected from a total of 32 infants. Three infants had incomplete neurodevelopmental measures or dropped out from the study, resulting in 14 infants in the control group and 15 infants in the intervention group (Table 1) at 12 months. There were no differences in socioeconomic status, smoking, prepregnancy body mass index, anxiety, and depressive symptoms between the mothers’ groups and no differences in anthropometric measurements or obstetric differences at birth between infants’ groups. At 12 months, the intervention group presented higher scores for gross motor and cognitive domains. In comparison to the whole sample, the subsample where cord blood was collected had a significantly higher number of boys (p = 0.01) and mothers presented higher scores on BAI (p = 0.04) (Suppl. File 1. Supplementary Table 1).
Table 1. Characteristics of mothers and their infants in control (n = 14) and intervention groups (n = 15)
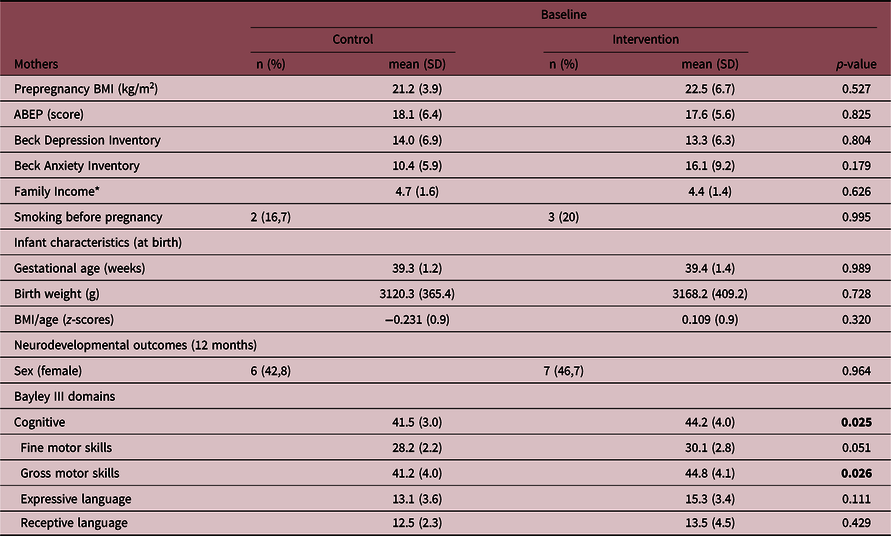
ABEP, Brazilian Association of Survey Companies; BMI, body mass index. We reinforce in bold the statistically significant differences (p-value < 0.05).
* Brazilian Minimal Wage.
Identification of methylation differences between intervention and control groups
By comparing DNAm levels for cord blood, intervention and control group had 3090 DMPs, being 1855 (60%) hypomethylated CpGs in the intervention group (Suppl. File 1. Suppl. Fig. 2). The 3090 DMPs were in 1267 genes, being 217 mapped within 1500 bp of the transcriptional start site (Suppl. File 2. Suppl. Table 3). Nineteen DMPs had delta-beta values > 20% (Suppl. File 1. Suppl. Table 3A).
The 3090 DMPs were tested for their potential association with the cognitive score of these infants at 12 months of age by multiple linear regression controlled by sex and using the cognitive domain score as outcome. Eighteen CpGs presented adj. R 2 > 0.4 (Suppl. Table 4), with 11 CpG mapped to the genes: C10orf118, C11orf58, C6orf174, GRIA1, LHFPL4, LOC100240726, MYOM2, RB1, STXBP6, TBL1XR1, and TOR1AIP2. From the CpGs not annotated to a gene, using ENCODE annotation, one CpGs (cg19610750) was located on a DNase hypersensitivity site annotated for multiple transcription factors, including CTCF.
We identified 21 DMRs, 15 (71%) hypomethylated in the intervention group. Fifteen DMRs were located at genes (HLA-DPB1, ABAT, ANKRD30B, PLEKHH3, ABCC13, LOC650226, LOC441666, PAX8, BOLL, RUFY1, LOC149837, HCG4P6, PF4, and TACSTD2), mostly located next to the transcriptional start site and/or promoter region of the respective gene (Suppl. Table 5). The DMRs were tested for their potential association with the cognitive score of these infants at 12 months of age by multiple linear regression controlled by sex using the cognitive domain score as outcome. PF4 was the unique DMR with a significant p-value (0.006) with a β (SE) = −5.48 (1.8) and R 2 adj = 0.30 associated with the cognitive outcome at 12 months (Suppl. Table 6).
Comethylation analysis of the 3090 DMPs pointed to six modules: MEblue (218 CpGs), MEbrown (138 CpGs), MEgreen (129 CpGs), MEred (68 CpGs), MEturquoise (219 CpGs) and MEyellow (136 CpGs). Annotation of CpGs as well as the respective modules (Suppl. File 3. Suppl. Table 6). MEturquoise (Z-summary = 7.42, MEcor = 0.13, p-value = 0.055) presented a low conservation score (Suppl. File 1. Suppl. Fig. 3). MEturquoise had 126 CpGs (out of 219) mapped to 96 genes. Four CpGs sites were classified as hub nodes in the MEturquoise module: one located at the body of CNTNAP1 and three CpGs located on the TSS200 of FILIP1.
Gestational age estimated by methylation point to a younger age of newborns receiving the intervention
Intervention and control groups did not present differences in the chronological gestational age at birth (Table 1; 39.4 × 39.3, intervention and control, respectively). The intervention group presented a smaller epigenetic gestational age mean compared to the control group (39.5 × 40.1 weeks, respectively) (p = 0.038) and AA (p = 0.025) (Fig. 1). Logistic regression controlled by sex and with gestational age as outcome variable indicated a coefficient of −0.35 towards intervention group (p = 0.016).
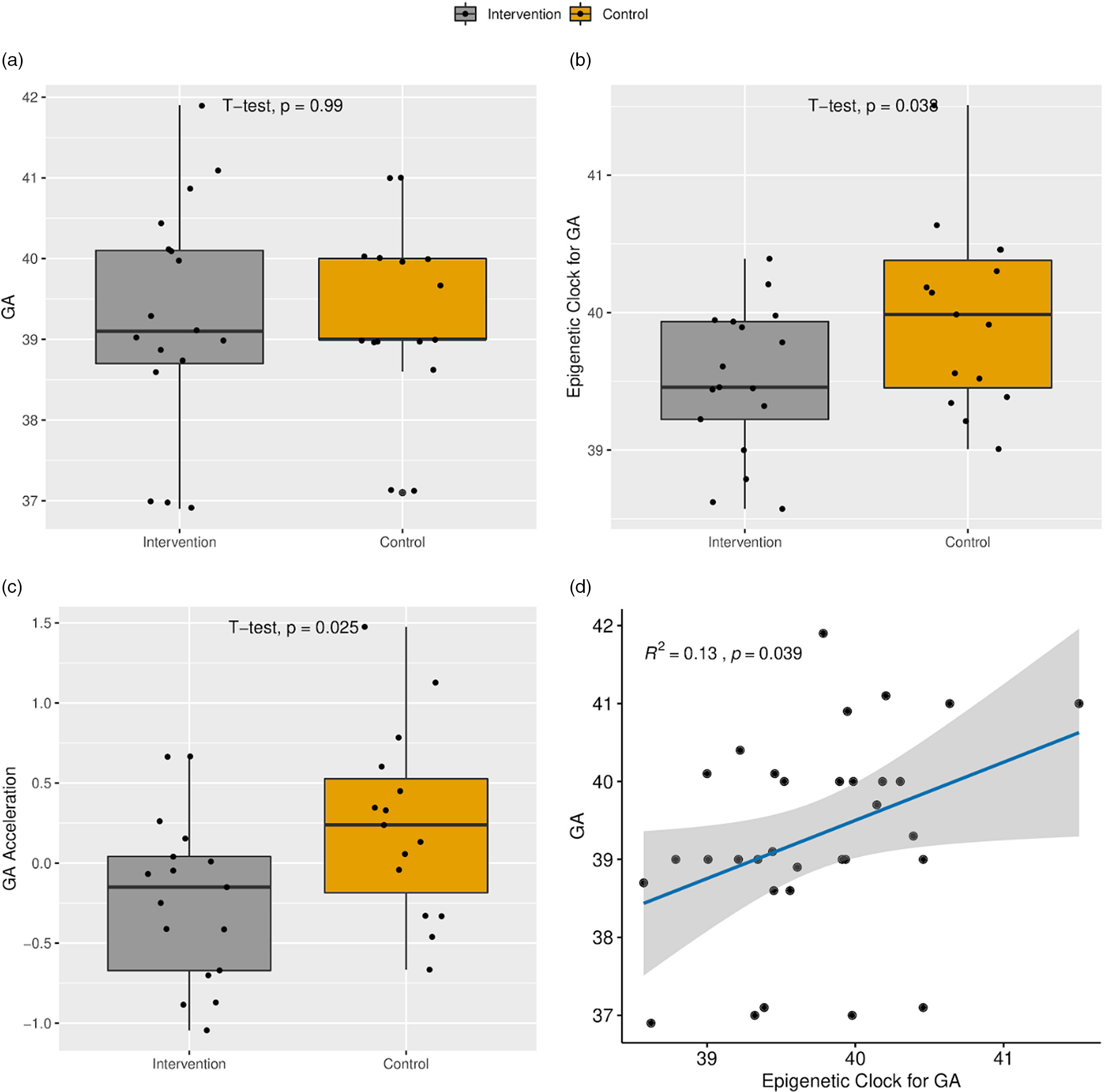
Fig. 1. (a) Boxplot of the chronological gestational age (GA) at delivery. (b) Boxplot of the epigenetic clock for GA. (c) Age acceleration. (d) Scatter plot of the relationship between epigenetic clock and GA.
DNAm show mediating effect on cognition at 12 months
Causal mediation analyses were applied to assess whether DNAm levels might be a mediator between intervention and infant’s cognition score at 12 months of age. (Suppl. File 1. Suppl. Table 7). Fig. 2 shows the results of CpGs selection pipeline to mediation analyses: (1) 18 CpGs associated with cognitive domain (adj. R 2 > 0.4), (2) 4 CpGs that were hubs in the less preserved comethylation module, (3) 21 DMRs, considering the averaged M-values of all CpGs within the region and (4) epigenetic clock as raw values.
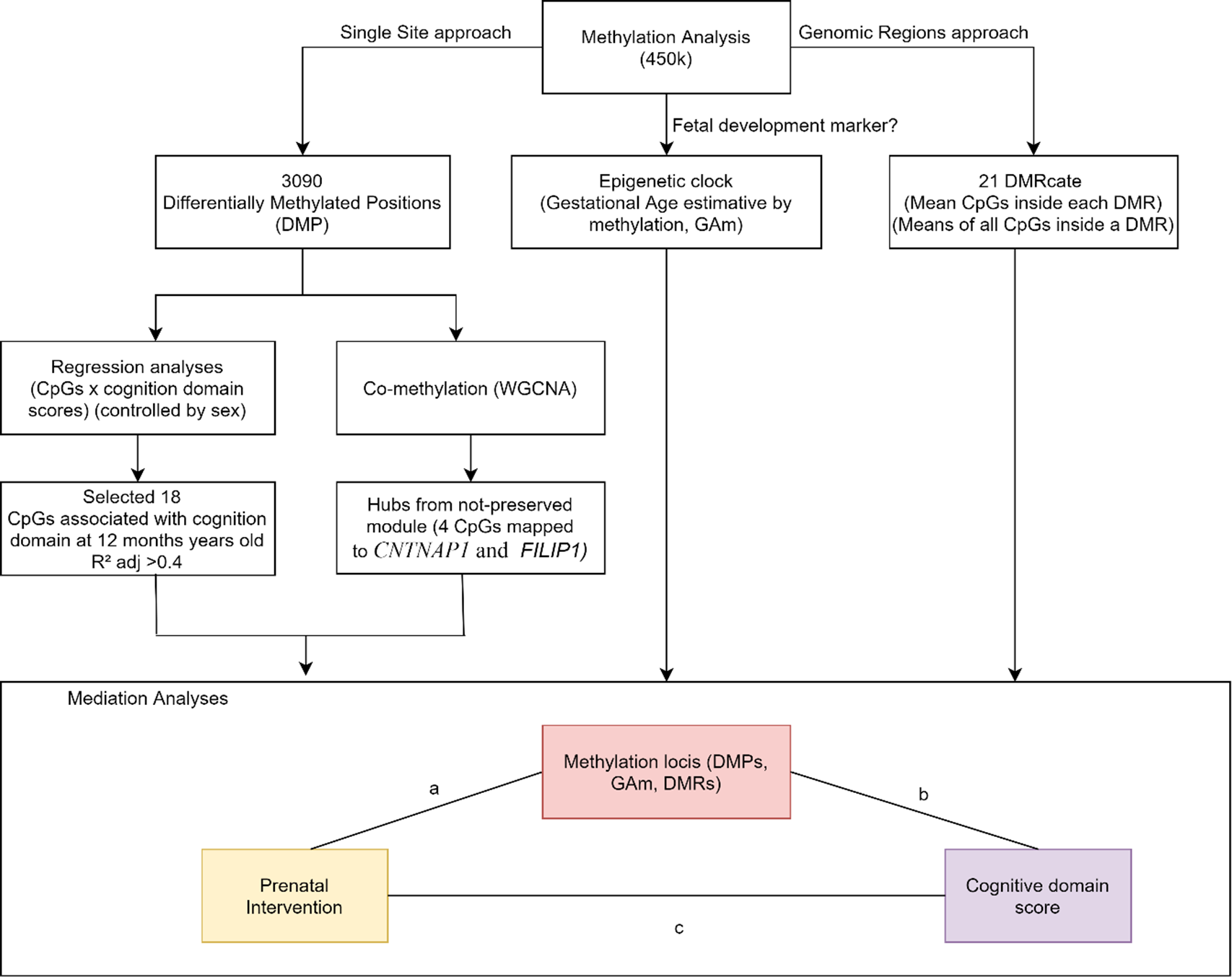
Fig. 2. DNAm variables approaches to select variables to be tested in causal mediation analysis. We tested the mediation effect of single sites (DMPs). DMRs and age acceleration in the relationship between the intervention and the cognitive score at 12 months of age.
A total of 18 DMPs were identified for significant ACME, with 11 located at TBL1XR, C11orf58, C6orf174, TOR1AIP2, MYOM2, C10orf118, STXBP6, RB1, LOC100240726, GRIA1, LHFPL4. However, there are inconsistencies in the signal for TBL1XR1 and GRIA1 that impact the total effect and make it impossible to talk about the proportion of the effect that was mediated. Moreover, three DMRs located at BOLL, LOC650226 and PF4 presented significant ACME, however, LOC650226 only with partial mediation (ADE significant). Additionally, we applied a sensitivity analysis to verify if there are no omitted variables that could bias the causal mediation results. The randomization of the intervention assures that no omitted variables will bias the exposure and DNAm, but since it is not possible to randomize the mediator, confounders could affect DNAm and cognition scores. Sensitivity analyses reinforced the causal mediation effect of DNAm (Suppl. File 4), suggesting if we have unknown confounders the mediation should be observed. The correlation between residuals (Rho) when indirect effects is zero (ACME = 0) as shown in Table 2. Our results presented a mediation of DNAm in the association between intervention and cognitive domain at 12 months of age (Table 2). Finally, the gestational age and AA given by the epigenetic clock did not show mediation effects.
Table 2. Mediation effects of DNA methylation in the relationship between intervention and at birth and cognitive scores at 12 months
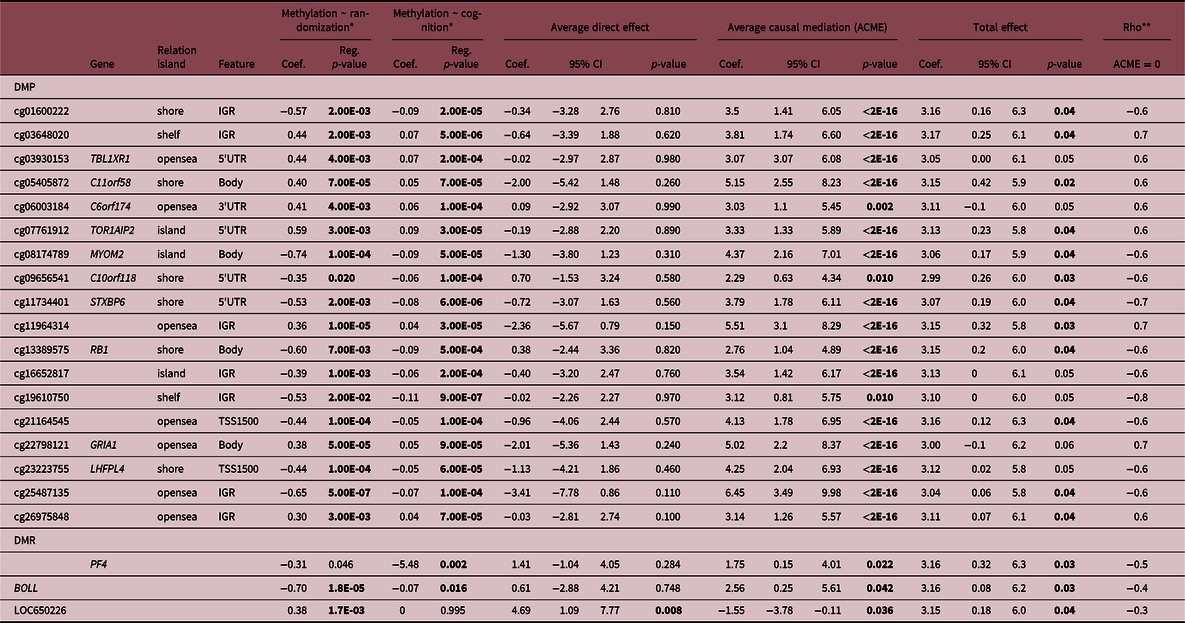
We reinforce in bold the statistically significant differences (p-value < 0.05).
* Controlled by sex.
** Sensitivity test: Rho at which ACME = 0.
Discussion
Addressing interventions prior to birth that could improve neurodevelopmental outcomes would be a significant advancement in modern mental health research. Thus, testing prenatal interventions to reduce stress for mothers living in volatile situations is important to determine the impact on brain development of the developing fetus. It is proposed that maternal stress and fetal development may be mediated by epigenetic mechanisms, including DNAm, of the offspring. In this study, we investigated cord blood DNAm differences (DMPs, DMRs, and hubs from comethylation network) and the epigenetic clock between intervention and control groups, that could act as mediators of the effects of a nurse-led home-visitation intervention program during pregnancy in child cognitive development at 12 months. Given that our sample size was limited, we only selected DMPs that could contribute to the cognition development at 12 months (DMPs explaining at least 40% of the outcome) to test mediation effect.
In terms of potential mediators, 5 out of 12 DMPs with a significant mediation effect (ACME and total effect) were mapped to genes involved in neuron functions and some associated to neurodevelopmental disorders: MYOM2 (cg08174789), RB1 (cg13389575), C10orf118 (cg05405872), STXBP6 (cg11734401), and TOR1AIP2 (cg07761912).Reference Boland, Nazor and Tran37–Reference Matsui, Nieto-Esteìvez, Kyrychenko, Schneider and Hsieh39 C10orf118 and STXBP6 are involved with vesicle trafficking and signaling Reference Scales, Hesser, Masuda and Scheller40 and are associated with dendritic protein localization in neurons. Reference Al-Bassam, Xu, Wandless and Arnold41 TOR1AIP1 encodes cofactors for the ATPase TorsinA, important to maintain developing neurons, during neuronal differentiation. Reference Rampello, Prophet and Schlieker42 The DMPs for these three genes (C10orf118, STXBP6, and TOR1AIP1) are located at the 5′UTR increasing the probability that their methylation is associated with downregulation of gene expression. Thus, we suggest that the cognitive score improvement by the intervention is mediated by a decrease in TORIAIP2 expression and an increase in the expression of C10orf118 and STXBP6, with the former mediating 70% of the total effect. Another interesting CpG that mediates the intervention effect on cognition is cg19610750, despite located at an intergenic region, it is annotated in binding sites for the transcriptional factors CTCF and EGR1, both involved with fetal development. Reference Strong, Butcher and Singhania43,Reference Duclot and Kabbaj44 We highlight the cg23223755, located at LHFPL4, a significant DMP also present in the MEturquoise, the least preserved module between cases and controls. It explained 40% of the association with infant’s cognition at 12 months. LHFPL4 forms a stable tripartite complex with ionotropic GABAA receptors (GABAARs) and neuroligin-2 in the brain. GABAARs mediate most of the fast-inhibitory transmissions in the brain. Moreover, LHFPL4 is required for the synaptic localization of GABAARs and inhibitory synaptic transmission in the hippocampus. Reference Yamasaki, Hoyos-Ramirez, Martenson, Morimoto-Tomita and Tomita45 We recognize that epigenetic changes in the umbilical cord blood do not necessarily directly reflect associated changes in the fetal brain. Nevertheless, surrogate tissue can be informative even if the investigated tissue is not directly involved in the phenotype of interest, as they might reflect the exposure to environmental influences as shown for the effects of prenatal sub nutrition, Reference Tobi, Slieker and Luijk9 chemical exposure and psychosocial adversity Reference Lyall, Schmidt and Hertz-Picciotto46 or combat. Reference Rutten, Vermetten and Vinkers47 Thus, although variation in peripheral tissue may not directly reflect epigenetic variation in the brain, they may still represent usable biomarkers as they reflect an exposure that leads to the phenotype in a different tissue. Reference Kumsta48
Considering the perspective that an acceleration of age measured by DNAm was previously related to both prenatal stress and measures of growth at birth, Reference Shiau, Wang and Liu49,Reference Suarez, Lahti and Czamara50 we were particularly interested if the epigenetic clock could mediate the relationship between intervention and cognition. Although not a mediator, in the present study, the intervention group presented a lower AA in comparison to the control group. In our sample only 1 mother from the intervention and 2 from the control groups smoked during gestation, and no significant different smoking status was observed between groups. Even though it is important to note that a previous study found AA differences associated to mothers that smoked during gestation compared to nonsmokers mothers. Reference Javed, Chen, Lin and Liang51 Epigenetic clock is an indirect measure of cellular senescence, previous findings have suggested that the aging process can begin even before birth. Reference Lewis, Cleal and Hanson52 These findings suggest that the intervention group has a protracted cellular senescence.
It is of great interest that the hypomethylated PF4 DMR was found to mediate 55% of the total effect on the association between the intervention and cognitive scores. PF4 encodes a member of the CXC chemokine family that initiates a signal transduction cascade of acute and delayed functions in human monocytes, including phagocytosis, respiratory burst, survival, and the secretion of cytokines. Reference Kasper, Brandt, Brandau and Petersen53 PF4 was a hub in a coexpression network analysis comparing individuals exposed to psychosocial stress early in life to controls. Reference Dieckmann, Cole and Kumsta54 The module was enriched for chemokine activity and for secretory vesicles functional biological processes. Moreover, PF4 overexpression contributes to an anti-aging effect in aged mice taking collagen peptides. Reference Song, Zhang, Luo, Zhang and Li55 We describe PF4 as hypomethylated in the intervention group, which could be associated with gene overexpression, and suggest that it could be contributing to the lower AA observed and the better cognitive scores in this group.
As with any study, there are some limitations that require further discussion. First, the sample size calculation of the randomized clinical trial was based on electroencephalography differences between cases and controls Reference Fracolli, de Reticena, de Abreu and Chiesa20 and not on methylation differences. Moreover, we used a subsample of the original RCT that reduced statistical power to detect potential differences in the variables studied. Although there were no differences of sex and the anxiety scale between groups in the subsample, compared to the RCT sample, the subsample with cord blood information had more boys (p = 0.01) and mothers presented higher scores on BAI (p = 0.04). Moreover, the postnatal intervention could also interfere in the outcome. Finally, the inclusion of first participants occurred previously to the clinical trial registration. We emphasize, however, that the registration happened before the end of the study. At the time we started the inclusion of the participants, we were not used to clinictrials.gov procedures.
In conclusion, we found that a maternal psychosocial intervention could impact on neurodevelopmental outcomes mediated by epigenetic mechanism. Future studies are encouraged to improve timing of measures and increase their sample size to best validate our findings.
Supplementary information
For supplementary material for this article, please visit https://doi.org/10.1017/S2040174421000738
Acknowledgments
We thank all study participants and the research team members for their valuable contributions.
Author contributions
VLVE wrote the paper, performed bioinformatic and mediation analysis; VDG performed bioinformatic analysis; ASF performed bioinformatic analysis; DJH contributed data analyses and co-wrote the paper; GG all laboratories procedures; HC supervised mediation analysis; AF-S cord blood collection; RPV cord blood collection; ECM performed clinical intervention and data collection; GVP performed clinical intervention and data collection; AC performed clinical intervention and data collection; LF performed clinical intervention and data collection; AM performed clinical intervention and data collection; AF performed clinical intervention and data collection; AA performed clinical intervention and data collection; MM contribute to methylation analyses and wrote the paper; HPB conceive the work, coordinated all the study, revised bioinformatics and mediation analysis, wrote the paper; All authors revised the manuscript.
Financial support
This work is supported by Grand Challenges Canada (GCC), Fundação Maria Cecília Souto Vidigal, Bill & Mellinda Gates Foundation, Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001 and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 2018/18560–6).
Consent for publication
This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
Competing interests
The authors declare that they have no competing interests.
Ethical standards
The present study was approved by the Ethics Committee of the University of São Paulo Medical School (ref: 052/15), the University Hospital of the University of São Paulo, and by the Sao Paulo Municipal Health Department. Participants and their caregivers provided informed consent. The study was registered at clinicaltrial.gov (NCT02807818).







