The influence of the gut microbiota on health and disease has become increasingly evident over the last decade( Reference Marchesi, Adams and Fava 1 ). The evolution in high-throughput sequencing has been critical in enabling the study of the microbiota. Indeed, several large collaborative studies have characterised distinct microbiota patterns relating to disease risk( Reference Wang, Hoenig and Malin 2 , Reference Qin, Li and Cai 3 ). In the context of the increasing prevalence of obesity and type 2 diabetes mellitus, it is striking that gut microbiota composition appears to be different in animals and humans with such diseases, compared with healthy controls( Reference Qin, Li and Cai 3 – Reference Ley, Turnbaugh and Klein 5 ). Thus, the gut microbiota has been identified as an important, potentially modifiable factor that contributes to conditions of metabolic dysfunction.
Dietary modulation of the gut microbiota composition using fermented products containing specific probiotics has been practiced for many years as part of nutritional therapy( Reference Collins and Gibson 6 ). Defined as ‘live micro-organisms that provide benefits to the host when administered in adequate quantities’( 7 ), probiotics have been extensively studied in relation to health promotion and disease prevention, both as additives to dairy products and as isolated bacterial strains( Reference Goldin and Gorbach 8 ). The efficacy of probiotics in promoting health remains disputed, and results differ depending on the outcome of interest, choice of the probiotic strain, formulation of the probiotic and duration of the intervention.
The use of probiotics to improve metabolic health is a relatively new indication for probiotic treatment. Of particular interest is the potential for probiotics to modulate inflammatory status, as demonstrated both in cell culture( Reference Kekkonen, Lummela and Karjalainen 9 ) and in some human studies( Reference Kekkonen, Lummela and Karjalainen 9 – Reference Asemi, Samimi and Tabassi 11 ). Chronic, ‘low-grade’ inflammation is widely recognised as playing a role in the pathological process leading to metabolic disease( Reference Gonzalez-Chavez, Elizondo-Argueta and Gutierrez-Reyes 12 ). Furthermore, inflammation in the case of metabolic disorders has been associated with changes of the intestinal microbiota that appear to be in part modulated by the gut-derived factor, lipopolysaccharide (LPS). Indeed, Cani et al. demonstrated that LPS can induce an inflammation that mimics inflammation that is induced by a high-fat meal( Reference Cani, Amar and Iglesias 13 ), and proposed that a high-fat diet could contribute to inflammation by increasing the transfer of LPS derived from the gut microbiota across the intestinal barrier( Reference Cani, Possemiers and Van de Wiele 14 ). The modulation of inflammatory status by dietary intervention, particularly during the early stages of metabolic dysfunction, could thus form a useful part of disease prevention.
Despite the potential for probiotics to modulate metabolic health by gut microbiota-related mechanisms, the gut microbiota is not always assessed in intervention studies that consider the effect of probiotics on metabolic health outcomes( Reference Asemi, Samimi and Tabassi 11 , Reference Bernini, Simao and Alfieri 15 , Reference Mohamadshahi, Veissi and Haidari 16 ), and in cases where analysis is completed, targeted or semi-targeted approaches are often adopted( Reference Kekkonen, Lummela and Karjalainen 9 , Reference Meyer, Elmadfa and Herbacek 10 ). The development of untargeted metagenomic techniques, such as 16S sequencing, offers a more comprehensive approach to understand how the gut microbiota might be influenced by these dietary interventions.
Milk can modulate both the inflammatory response( Reference Bordoni, Danesi and Dardevet 17 ) and the composition of the gut microbiota( Reference Charbonneau, O’Donnell and Blanton 18 ). As dairy products are important vectors for the delivery of probiotics to humans, in the present study, we explore the impact of two dairy product dietary interventions on metabolic and inflammatory outcomes, with parallel evaluation of the faecal microbiota dynamics. First, we test the hypothesis that a probiotic yogurt, compared with milk acidified with glucono-δ-lactone, can reduce transient inflammation induced by a high-fat meal challenge. The validated test models a metabolic stimulus which aims to mimic the inflammatory stress that precedes metabolic dysfunction( Reference Schwander, Kopf-Bolanz and Buri 19 ). Second, we test the hypothesis that these interventions alter the gut microbial composition.
Methods
Subjects
A total of fourteen healthy young men were recruited by a poster campaign (December 2013–March 2014). Inclusion criteria were applied to select subjects aged 18–40 years with a stable, healthy BMI (18·5–25·0 kg/m2), regular dietary and physical activity habits, and no evidence of dietary intolerances, restrictions or adverse reactions to dairy products. Exclusion criteria included chronic or acute disease, hypertension, regular medication, moderate or intense physical activity (exceeding 6 h/week), nutritional supplements, antibiotic treatment in the 6 months preceding the study and a history of anaemia. These criteria were verified during an inclusion visit that included a physical medical examination, dietary and physical activity assessments, standard anthropometrics and bioimpedance analysis (ImpDF50; ImpediMed). Fasting glycaemia, insulinaemia, lipid profile, full blood count and Fe profile were evaluated. This study was conducted according to the guidelines laid down in the Declaration of Helsinki and all procedures involving human subjects were approved by the regional committee for human experimentation (approval no.: 392/13, Vaud, Switzerland). Written informed consent was obtained from all subjects. The trial was registered at clinicaltrial.gov (registration number: NCT02230345).
Interventions
Two dairy products were tested during the study: yogurt containing the probiotic Lactobacillus rhamnosus GG (LGG) (ATCC 53103) and milk acidified with d-(+)-glucono-δ-lactone (2 %) in order to mimic texture, pH and physical properties of the yogurt. All dairy products provided during the study were derived from the same batch of full-fat homogenised, ultra-high-temperature-treated milk (3·5 %) that was supplied by Emmi AG. The probiotic yogurt was prepared by fermentation using Lactobacillus delbrueckii spp. bulgaricus and Streptococcus thermophilus (Thermophilic Yoflex® culture; Chr. Hansen) (online Supplementary Methods). Bacterial counts were carried out for the production batches to confirm a minimum of 1·00×106 colony-forming units (CFU)/g per strain: L. delbrueckii spp. bulgaricus 9·04×107 (sd 3·55×107) CFU/g, S. thermophilus 6·50×108 (sd 1·04×108) CFU/g and LGG 2·83×106 (sd 6·53×105) CFU/g (online Supplementary Fig. S1). Participants consumed dairy products that were between 4 and 15 d post-production. The nutritional composition of the products is detailed in the online Supplementary Table S1.
Experimental design
The experimental design followed the structure of a randomised, double-blind, cross-over trial (Fig. 1(a)). This procedure was used to evaluate the postprandial and short-term effects of the two dairy products investigated, probiotic yogurt and acidified milk. The dairy products were randomly allocated to volunteers (seven volunteers per test sequence). Each product was tested over a 2-week intervention phase.

Fig. 1 Overview of the study design. (a) Participants were assigned randomly to group 1 or group 2 to test the probiotic yogurt and acidified milk in a cross-over design. Wash-out periods followed each test phase and a run-in preceded the beginning of the study. (b) Metabolic and inflammatory assessments. ![]() , Evaluations on dairy product test;
, Evaluations on dairy product test; ![]() , evaluations on high-fat meal test; CD1–5, control diet 1–5; HFM1–3, high-fat meal test days 1–3; D1–2, dairy product test day 1–2; FS1–8, faecal samples 1–8; HOMA, homoeostatic model assessment; hsCRP, high-sensitivity C-reactive protein; CCL2, chemokine ligand 2; CCL5, chemokine ligand 5.
, evaluations on high-fat meal test; CD1–5, control diet 1–5; HFM1–3, high-fat meal test days 1–3; D1–2, dairy product test day 1–2; FS1–8, faecal samples 1–8; HOMA, homoeostatic model assessment; hsCRP, high-sensitivity C-reactive protein; CCL2, chemokine ligand 2; CCL5, chemokine ligand 5.
Two distinct types of postprandial tests were carried out during the study: (1) dairy product test (D1, D2) and (2) high-fat meal test (HFM) (HFM1, HFM2 and HFM3). The first type of test evaluated the postprandial response to a single dose of the assigned dairy product (800 g of probiotic yogurt or acidified milk). This was conducted on the first day of each test phase. The test marked the beginning of the daily intake phase that required a daily consumption of 400 g (as per Swiss guidelines for recommended dairy product intake( Reference Infanger 20 )) of the assigned dairy product for 2 weeks. At the end of this period, a second type of postprandial test was carried out to assess whether the daily intake of probiotic yogurt or acidified milk could influence the response to a meal that normally induces a state of transient inflammation. This test used a mixed-meal challenge (adapted from Schwander et al. ( Reference Schwander, Kopf-Bolanz and Buri 19 )) formulated to contain an elevated quantity of energy in the form of saturated fat and sugar, and no dairy products (Table 1). The baseline measurements for this test were completed during a run-in period that preceded the first intervention (HFM1 in Fig. 1(a)). During this 4-week run-in period, a fixed dose (400 ml/d) of full-fat milk was provided to the volunteers to normalise the background dairy product intake before the two dairy product interventions. The same conditions were applied during two 3-week wash-out phases that followed each intervention period.
Table 1 Composition of the high-fat meal test (HFM) used to induce postprandial inflammationFootnote *
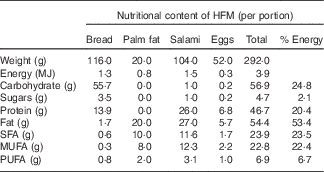
* Adapted from Schwander et al.( Reference Schwander, Kopf-Bolanz and Buri 19 ).
Dietary and lifestyle restrictions
Dietary restrictions and assessments were completed during all study phases with additional monitoring of physical activity. Participants were asked to maintain a stable level of physical activity, comparable with their usual pattern of activity, during the study and to avoid intense physical activity during the 3 d preceding each test day. This was evaluated by ActiGraph wGT3X accelerometers (ActiGraph) worn on the waist during five phases of the study: run-in, test phases (1, 2) wash-out phases (1, 2). Accelerometers recorded data in 60 s epochs.
Dietary intake was semi-controlled during the study. Dietary restrictions excluded all dairy products not provided by the study organisers and included specific guidance on portions of fermented foods, alcohol intake and caffeine intake to replicate normal baseline eating patterns. During each test phase of the study, 3-d self-completed dietary records (including 1 weekend day) were completed. In addition, participants self-reported compliance with the assigned dairy product using a daily record. Before each of the five test days, participants followed a 3-d controlled diet (55 % carbohydrate, 30 % fat, 15 % protein – an example menu is available in the online Supplementary Table S2) (Fig. 1(a), control diet 1–5 (CD1–CD5)). All foods were provided by the study organisers but consumed under free-living conditions. Portions were adapted to meet individual nutritional requirements (estimations completed using the Harris–Benedict equations and physical activity levels 1·5–1·7). Product tolerance and well-being was assessed during each test phase intervention.
Study days
All five test days were completed at the Centre of Clinical Research, Lausanne, Switzerland. On each test day, the participants arrived in a fasted state and a standard clinical assessment that included body weight and composition was carried out (bioimpedance analysis with ImpDF50). A Venflon catheter was placed laterally in the arm of the participant, and after a resting period of 20 min, the first fasting blood sample was taken. The volunteer was then given the test meal, and 6-h postprandial sampling was initiated. A visual analogue scale questionnaire (based on the work of Flint et al.( Reference Flint, Raben and Blundell 21 )) was used to assess satiation in the fasted state and postprandially until test completion.
Sampling
Blood samples were taken at selected time points during the test days (Fig. 1(b)). No blood sampling was done after the completion of wash-out 2. Serum, plasma and whole-blood samples (PAXgene® collection tubes; Qiagen) were prepared according to standard protocols for the selected biomarker. Samples for LPS endotoxin analysis were prepared in sterile tubes (0·2 % heparin). All blood samples were stored at –80°C.
A total of eight faecal samples were collected from the participants during the study (Fig. 1(a)). These samples were processed under sterile conditions within 4 h of sample collection. Aliquots (200 mg) were added to 2 ml of glycerol–brain heart infusion solution (100 ml glycerol, 37 g Brain Heart Solution, 1 litre distilled water). Following homogenisation by agitation (13 g , 10 min), samples were stored at –80°C. Samples were washed three times with PBS (centrifugation 16 000 g , 2 min) and suspended at 95°C before DNA extraction, which was performed using the QIAamp Fast DNA Stool Mini Kit (Qiagen). Urine samples were collected as a fasted ‘spot test’ and as a single pooled sample for the 6 h of each test day using a tube with no additives.
Metabolic and inflammatory marker analyses
Classical parameters of metabolic health and selected circulating inflammatory markers were analysed at selected time points on the test days (Fig. 1(b)). The COBAS® 8000 platform (Roche Diagnostics International AG) was used to assess routine biomarkers (insulin was assessed by the electro-chemiluminescence immunoassay assay, all other biomarkers were measured as previously described by van Leckwyck et al. ( Reference van Leckwyck, Kong and Burton 22 )). NEFA were assessed using Wako reagents for NEFA analysis (Wako Diagnostics) on the Pentra 400 platform (ABX; Horiba). Endotoxin levels were assayed using QCL-1000 LAL endpoint assay (Lonza). IL6 and TNFα concentrations were measured by the Bio-Plex ProTM Human Cytokine Standard 27-plex, Group 1 assay (BioRad)( Reference Schmid, Petry and Walther 23 ) (intra-assay CV 0·52 % for TNFα, 0·48 % for IL6; inter-assay CV 17 % for IL6, 8 % for TNFα). Chemokine ligand 2 (CCL2) and chemokine ligand 5 (CCL5) concentrations were measured with the Bio-Plex ProTM Human Cancer Biomarker Panel 2, 2-plex assay (BioRad) (intra-assay CV 1·5 % for CCL2, 1·1 % for CCL5; inter-assay CV 23 % for CCL2, 24 % for CCL5). Both assays were completed using the Luminex® MAGPIX® system (Luminex® Cooperation) that applies magnetic bead methodology. All kits for cytokine and chemokine assessments were from the same kit production batch and all samples from the same volunteer were analysed on the same plate to limit the effect of inter-assay variability. These analyses were performed according to the manufacturer’s instructions.
Analysis of faecal microbiota
DNA libraries were prepared with the extracted faecal DNA using the 16S Metagenomic Sequencing Library protocol, as defined by the Illumina MiSeq System. Primers S-D-Bact-0341-b-S-17 and S-D-Bact-0785-a-A-21 were chosen to target the V3–V4 regions of the bacterial genome( Reference Klindworth, Pruesse and Schweer 24 ). Verification of the library quality was completed using the Fragment Analyzer (Advanced Analytical). Sequencing was completed on the Illumina MiSeq and read-quality analysis assessed on the Illumina BaseSpace platform.
Nutritional and physical activity analyses
Accelerometry data were analysed with ActiLife 6 (version 6.10.0). Wear-time validation was completed using the Choi algorithm( Reference Choi, Liu and Matthews 25 ). Data were included in the analysis if the total wear-time for the day assessed exceeded 8 h( Reference Evenson and Terry 26 , Reference Chinapaw, de Niet and Verloigne 27 ). Outcome parameters were total vector magnitude counts, average vector magnitude counts, counts in light, moderate, vigorous, very vigorous and combined moderate-vigorous activities (Freedson Adult VM3 reference cut-off points( Reference Freedson, Melanson and Sirard 28 )), total number of bouts, total counts during bouts, sedentary bouts, activity energy expenditure, metabolic equivalent of task( Reference Sasaki, John and Freedson 29 ), and wear-time.
Food-intake records were analysed by a registered dietitian using the Nutrilog software, version 2.70 with the databases OSAV 5.1( 30 ), Ciqual( 31 ), Aliments de Marques ( Reference Blanc 32 ) and manual entry of nutritional data, if available. Total energy intake and macronutrient intake were assessed for all test periods.
Bioinformatic data processing
The raw paired-reads were assembled into contigs using Pandaseq software( Reference Masella, Bartram and Truszkowski 33 ). Only contigs without any ambigous nucleotides and with a length between 390 and 450 nucleotides were retained in the subsequent analysis. Reference operational taxonomic unit (OTU) sequences were selected using the USEARCH pipeline( Reference Edgar 34 ) using the full data set after discarding sequences that were not repeated at least ten times across all samples. Three sequences related to yogurt strains were manually added to the set of OTU reference sequences: Streptococcus salivarius spp. thermophilus, L. delbrueckii spp. lactis and Lactobacillus casei/paracasei. The final abundance of OTU in each sample was counted using the USEARCH package( Reference Edgar 34 ).
The 16S rRNA sequences together with taxonomic annotations were downloaded from SILVA database (version 123)( Reference Quast, Pruesse and Yilmaz 35 ). All sequences with a pintail quality score( Reference Ashelford, Chuzhanova and Fry 36 ) <0·9 were removed from the database. The reference sequences for OTU were mapped against the database and taxonomy was assigned to the OTU on the basis of best hits, ensuring that the obtained nucleotide similarity exceeded 97 %. The species level assignment was kept only in cases of sequences that displayed similarity exceeding 99 %. Any sequence that was unassigned using this method was further subjected to classification using online SINA alignment service with default parameters( Reference Pruesse, Peplies and Glockner 37 ).
Statistical analyses
The study was designed with two objectives: (1) to explore the changes of metabolomic parameters during postprandial dairy product tests and (2) to study the impact of short-term dairy intake on inflammation. The sample number was chosen to reflect the exploratory nature of the first objective of the study within a highly controlled study design. We report here the results of our second objective. The desired sample number could not be determined because of the absence of previous clinical studies with a similar intervention.
All statistical analyses were performed using R (version 3.2.4)( 38 ) with applied packages MESS (version 0.3–2)( Reference Ekstrøm 39 ), DESeq2 (version 1.10.1)( Reference Love, Huber and Anders 40 ), phyloseq (version 1.14.0)( Reference McMurdie and Holmes 41 ) and dendextend (version 1.1.8)( Reference Galili 42 ). The Wilcoxon signed-rank test was used to compare the effect of the two dairy product interventions on fasting biomarkers and on the postprandial response to the HFM. Differences were considered significant at P≤0·05. Fasting assessments were evaluated by calculating the respective change after each intervention compared with the baseline levels at the beginning of each test phase. Linear evaluation of the incremental AUC was completed to assess the postprandial response to the HFM (MESS package( Reference Ekstrøm 39 )). Missing data points for the postprandial response were treated by extrapolation of the postprandial curve where possible, or data were removed if the missing data concerned fasting time points. A pre-test was completed to confirm the assumption of negligible carryover effects, as described by Wellek & Blettner( Reference Wellek and Blettner 43 ). The treatment effect was then assessed by calculating within-subject differences for the two sequence groups. The response to the HFM after the dairy product administration was also compared with the baseline response to this test. For this evaluation, the response to each dairy product was compared separately with the baseline test using pairwise comparisons for all subjects pooled, given that all baseline tests preceded the dairy product interventions and no carryover effects were observed. Dietary intake and physical activity changes were assessed using the Kruskal–Wallis rank-sum test using P≤0·05 to define significant differences.
Microbiota analysis was completed on taxa present at a minimum mean abundance of 0·01 %/volunteer, in at least three volunteers. Cluster analysis was completed at the species level using Spearman’s correlation (amap, version 0.8–1.4( Reference Lucas 44 )). DESeq2 was used to complete differential analyses on the microbiota using the Wald test and with significance assessed with P adj≤0·05( Reference Love, Huber and Anders 40 ) (Benjamini–Hochberg correction( Reference Benjamini and Hochberg 45 )). The microbiota composition after each test phase was compared directly and analysis was also completed separately for each intervention phase with comparison with pooled samples from all normal-milk phases (run-in, wash-out 1 and wash-out 2). Carryover effects of the intervention were assessed by comparison of samples from wash-out phases after the respective interventions. Time-course changes were assessed by pairwise comparison of each normal-milk phase. In view of the significant inter-individual variation, analysis was completed using samples from volunteers who provided at least one viable faecal sample for each condition being compared in the differential analysis. Diversity indices were assessed by Phyloseq( Reference McMurdie and Holmes 41 ), using the Shanon and Simpson indices.
Results
Participant characteristics
A total of fourteen healthy young men were enrolled in the study and randomly allocated to one of the two test sequences (group 1: acidified milk–probiotic yogurt; group 2: probiotic yogurt–acidified milk) (online Supplementary Fig. S2). One subject from group 1 was excluded from all analysis because of suspected non-compliance with dietary restrictions that was detected during the microbiota analysis. High levels of the OTU for L. casei and L. paracasei were detected for this volunteer during the second wash-out phase (faecal sample 7) (online Supplementary Fig. S3(J)). These two bacterial strains are widely used in cheese fermentation and added to dairy products for their probiotic qualities( Reference Carafa, Nardin and Larcher 46 , Reference Holzapfel, Haberer and Geisen 47 ). The distinct spike in the number of reads apparent for faecal sample 7, together with the absence of the bacteria in all other conditions, is consistent with a discrete intake of a prohibited fermented or probiotic food. Furthermore, negligible counts for the three probiotic yogurt strains were detected for this volunteer after the consumption of the probiotic yogurt (online Supplementary Fig. S3(B, G)). The absence of bacterial strains for all other volunteers during run-in, wash-out and acidified milk phases (online Supplementary Fig. S3 (A, C–F, H, I)) indicate compliance with the dairy product dietary restrictions. A second subject (group 2) did not complete the final visit (post-acidified milk) because of acute illness. All the analysis concerning probiotic yogurt was thus completed with twelve participants, whereas the analysis of acidified milk was completed with thirteen participants. Baseline clinical parameters were all within the respective reference ranges (Table 2) and showed no difference between the two groups, except for BMI, which was significantly higher for sequence group 1.
Table 2 Participant characteristics (Medians and interquartile ranges (IQR))

DBP, diastolic blood pressure; SBP, systolic blood pressure; HOMA, homoeostatic model assessment; HbA1c, glycosylated Hb, type A1c.
Dietary and physical activity analyses
Dietary intake during the different phases of the study was not significantly different for macronutrients (P>0·05) (online Supplementary Table S3). In addition, energy expenditure assessed by Actigraph accelerometers showed no significant changes between test phases for sedentary, low, moderate-vigorous activity, or for total accelerometer counts by test phase (P>0·05); however, a trend of higher levels of moderate-vigorous as well as total counts was observed during the run-in phase (online Supplementary Table S4).
Fasting analyses
No changes in fasting metabolic and inflammatory markers were observed after 2 weeks’ probiotic yogurt intake compared with acidified milk intake, except for CCL2, which showed a greater increase after acidified milk intake (P=0·01) (online Supplementary Table S5). The cross-over analysis of the change in fasting markers suggested a difference between the volunteers in sequence groups 1 and 2 for insulin; however, no intervention effect was observed for this marker. In addition, no significant changes were observed for anthropometric measurements or body composition (data not shown).
High-fat meal test response
No significant differences were observed after probiotic yogurt intake compared with after acidified milk intake for any of the inflammatory and metabolic biomarkers used to assess the postprandial response to the HFM. However, the responses for IL6, CCL5 and TNFα to the HFM were significantly reduced after both interventions in comparison with the baseline evaluations that were completed during the run-in phase (P<0·001) (Fig. 2). A negative outlier was observed for all three biomarkers due to a raised fasting level of inflammation for one volunteer; however, given the use of non-parametric statistical tests, these data did not significantly alter the results. The maximum response for these biomarkers during the baseline (HFM1) test was observed between 90 and 120 min (online Supplementary Fig. S4). The postprandial response for inflammatory markers high-sensitivity C-reactive protein (hsCRP) and LPS, and metabolic biomarkers were not significantly different after probiotic yogurt or acidified milk intakes, compared with baseline assessments (data not shown).

Fig. 2 Postprandial response for inflammatory markers, (a) TNFα, (b) IL6 and (c) chemokine ligand 5 (CCL5), to the high-fat meal test at baseline and after the 2-week intake of acidified milk or probiotic yogurt. iAUC, incremental AUC. * P<0·001.
Microbiota
Cluster analysis of the microbiota data revealed a strong inter-individual variability at the OTU level, with all samples from the same volunteer being clustered together without exception (online Supplementary Fig. S5). Similar grouping was observed at the species and genus levels (data not shown).
The abundance of several bacteria was significantly different after the probiotic yogurt intervention compared with after the acidified milk intervention (online Supplementary Table S6). Notably, the two strains used in the fermentation process, S. salivarius spp. thermophilus and L. delbrueckii spp. bulgaricus, were both significantly more abundant after probiotic yogurt intake (log 2-fold-change (FC)=7·6 and 3·6, respectively, P adj<0·01). The strain Intestinibacter bartlettii was also more abundant after probiotic yogurt intake than after acidified milk intake (FC=2·1, P adj<0·01). Conversely, the Bifidobacterium kashiwanohense or Bifidobacterium pseudocatenulatum strains and the Megasphaera genus were less abundant after probiotic yogurt intake than after acidified milk intake (FC=–1·7, P adj<0·01 and FC=–1·7, respectively, P adj=0·03). These differences were consistently observed at the different taxonomic levels analysed.
Analysis of each intervention phase separately with respect to the baseline (normal milk) phases confirmed the specificity of the differences observed between the interventions (online Supplementary Tables S7 and S8). Transient increases in the relative abundance of S. salivarius spp. thermophilus and L. delbrueckii spp. bulgaricus were observed specifically after probiotic yogurt compared with baseline assessments (FC=6·0 and 7·0, respectively, P adj.<0·001) with no changes in the abundance of these strains after acidified milk intake compared with baseline (Fig. 3(a) and (b)). A non-significant increase in abundance was observed for the probiotic strain LGG after probiotic yogurt intake but not after acidified milk intake (Fig. 3(c)). The difference in abundance of B. kashiwanohense or B. pseudocatenulatum between the two interventions was shown to represent, specifically, a relative increase in the strains after acidified milk intake compared with baseline phases (Fig. 3(e)) (FC=1·4, P adj=0·04), with significant effects visible at all taxonomic levels evaluated. These species were not modulated by probiotic yogurt intake compared with baseline phases. The change in abundance of I. bartlettii was also associated with the acidified milk intake rather than the probiotic intervention, with significant differences in the abundance of the OTU for the strain after acidified milk intake, compared with baseline assessments (FC=–1·7, P=0·04).

Fig. 3 Changes in selected bacterial species after intake of probiotic yogurt and acidified milk: (a) Lactobacillus delbrueckii spp. bulgaricus; (b) Streptococcus salivarius spp. thermophilus; (c) Lactobacilllus rhamnosus GG; (d) Bilophila wadsworthia; (e) Bifidobacteria kashiwanohense/bifidobacteria pseudocatenulatum. Baseline milk (control) compared with interventions acidified milk and probiotic yogurt by differential analysis. * P adj≤0·05.
In addition, this analysis revealed further modulation of bacteria by the acidified dairy product interventions. Notably, significant reductions in abundance for the species Bilophila wadsworthia were observed after both acidified milk and probiotic yogurt intakes, with respect to baseline assessments (FC=–1·5, P adj=0·05 after acidified milk intake; FC=–1·3, P adj=0·03 after probiotic yogurt intake) (Fig. 3(d)). These changes were observed at all levels of taxonomic assessments up to and including class level. Specific changes in the abundance of the species Haemophilus parainfluenzae and six genera were observed after probiotic yogurt intake compared with baseline phases. Acidified milk was associated with relative reductions in the abundance of four OTU of unknown species, and relative increases in abundance of two OTU of unknown species (classified at the genus level as Ruminococcaceae UCG-014 and Lachnospiraceae UCG-004) compared with baseline phases.
Carryover analysis showed no significant changes of any taxa when comparing the wash-out after acidified milk intake with that completed after probiotic yogurt intake (data not shown). However, comparisons of the wash-out phases in study chronological order showed significant changes for the Clostridiaceae Family XIII AD3011 group that was also observed at higher taxonomic classifications from family to class level (online Supplementary Table S9)). Differences in bacteria were also observed in comparisons between the run-in phase and the two wash-out phases. Two OTU were different between the run-in and wash-out phase 1, and a further two OTU were different between the run-in and wash-out phase 2 (online Supplementary Tables S10 and S11).
Discussion
In this cross-over study, probiotic yogurt, compared with acidified milk, did not significantly modulate the postprandial inflammatory response associated with a HFM; however, both products similarly and significantly reduced this inflammatory response, compared with baseline assessments, in our relatively small cohort of healthy young men. These effects were observed in parallel to distinct changes in taxa of the gut microbiota including Lactobacillus, Streptococcus, Bifidobacterium, Bilophila and Ruminococcaceae. The modulation of the microbiota during the acidified milk intervention could be a mechanism for the unexpected effects of this dairy product on the inflammatory response.
We explored postprandial inflammation using a HFM model and observed that daily intake of probiotic yogurt and acidified milk similarly moderate this response. The use of ‘challenge’ tests such as our HFM can unveil metabolic adaptations that are not detectable in the fasted state. The model simulates physiological exposures to excessive energetic loads and consequentially aims to approach the reality of how the response to dietary excess can be modulated by other dietary components. Although such tests have been used in acute dietary interventions( Reference Edirisinghe, Banaszewski and Cappozzo 48 , Reference Burton-Freeman, Talbot and Park 49 ), few studies have applied the test after a longer exposure to a dietary stimulus( Reference Ellis, Edirisinghe and Kappagoda 50 ). Our findings confirm the usefulness of such an approach for evaluating inflammation associated with diet in a healthy population. One of the challenges of evaluating the inflammatory response in clinical studies is the absence of universally accepted biomarkers( Reference Calder, Ahluwalia and Albers 51 ). Using a selection of sensitive blood biomarkers, we detected changes in inflammatory parameters. As in the case of all biomarkers, the clinical significance of these findings needs to be further validated with clinical outcomes. Nevertheless, it was remarkable that we observed a consistent modulation of the IL6, TNFα and CCL5 responses to the HFM by the probiotic yogurt and acidified milk, with respect to baseline tests. Little response to the HFM was observed for hsCRP or LPS both for the baseline tests and tests after the intervention. This may relate to the sensitivity of the markers or could reflect the ‘metabolic flexibility’ of the healthy volunteers, which seeks to maintain homoeostatic control( Reference Schwander, Kopf-Bolanz and Buri 19 ).
The role of dairy products in inflammation is controversial, with evidence for both positive and negative effects described in a recent review of fifty-two clinical trials( Reference Bordoni, Danesi and Dardevet 17 ). Overall, a protective role of dairy products on inflammation was suggested in this review, although it was noted that dairy product characteristics and the health status of the population studied influenced this relationship. Given that our baseline challenge test was completed after a run-in period during which normal milk was consumed, it cannot be excluded that the apparent benefits we observe rather represent a relative reduction in inflammation, compared with stimulation of inflammatory parameters by normal milk. Unfortunately, the absence of blood sampling before all milk interventions or following the reintroduction of the participants’ normal diets prevented us from testing this hypothesis. Nevertheless, in view of the nutritional qualities of bovine milk, the distinct effects of both probiotic yogurt and acidified milk on inflammation remain an interesting finding.
The concept of modulating the intestinal microbiota in a targeted manner is important in view of the wide number of diseases that are associated with dysbiosis of the intestinal microbiota( Reference Carding, Verbeke and Vipond 52 ). In the current study, we concur with existing research that demonstrates the highly individual nature of the microbiota composition( Reference Turnbaugh, Hamady and Yatsunenko 53 ) and the relative stability of this identity over time( Reference Faith, Guruge and Charbonneau 54 ). Indeed, in previous work with salivary microbiota samples, we have already evoked the notion of the microbiota composition as a novel fingerprint which can differentiate individuals( Reference Leake, Pagni and Falquet 55 ). The stability of this identity is supported by a recent study that examined the microbiota changes following an antibiotic intervention( Reference Dethlefsen and Relman 56 ). Despite a rapid perturbation of the microbiota following antibiotic treatment, the inter-individual variability was still largely present following the intervention. Similarly, we observe a significant but distinct modulation of different taxa during the dairy product interventions, whereas the individual identity of the microbiota is maintained. Notably, the acidified milk intervention is associated with a significant increase in Bifidobacterium sp. OTU, which attests to the previously described function of gluconic acid as a prebiotic ingredient( Reference Asano, Yuasa and Kunugita 57 , Reference Tsukahara, Koyama and Okada 58 ). Whereas gluconic acid has already been used in isolation to induce an increase in this species( Reference Asano, Yuasa and Kunugita 57 ), this is the first study to demonstrate this quality in a dairy product formulation. Bifidobacteria have been associated with multiple health benefits, including benefits on metabolic and inflammatory parameters( Reference Bernini, Simao and Alfieri 15 , Reference Meng, Ba and Lee 59 ). The use of gluconic acid, transformed from glucono-δ-lactone, in combination with a probiotic dairy product, could be an interesting symbiotic approach to maximise the action of the probiotic.
The impact of our probiotic yogurt on the gut microbiota was highly specific to the strains that were present in the product, with limited effect on the commensal microbiota. These findings are in agreement with previous studies that investigated probiotics and in which the gut microbiota was studied( Reference Eloe-Fadrosh, Brady and Crabtree 60 ). The relatively low abundance of LGG in the probiotic yogurt culture explains the absence of significant increases of this strain after the probiotic yogurt test phase, despite the inoculation count exceeding that which has been previously established in dairy products as adequate to ensure faecal recovery of the probiotic( Reference Saxelin 61 ). One reason for this lower dosage was that at higher inoculation, LGG showed a strong inhibitory effect on the fermenting yogurt strain, L. delbrueckii spp. bulgaricus. Given the beneficial qualities of the yogurt fermenting strains, we opted for a compromise that would favour the growth of the fermenting strains while also meeting minimum levels of LGG to ensure faecal recovery. It should thus be noted that the effects that we observe on inflammation are the combination of a mixed culture including fermenting bacterial strains and LGG. The role of probiotic interventions in metabolic dysfunction is not clear, with an absence of effects reported in some studies( Reference Asemi, Samimi and Tabassi 11 , Reference Gobel, Larsen and Jakobsen 62 , Reference Mazloom, Hejazi and Dabbaghmanesh 63 ), although there is some evidence for a role of LGG in reducing inflammation( Reference Kekkonen, Lummela and Karjalainen 9 , Reference Schultz, Linde and Lehn 64 ) and even a report for the role of conventional yogurt in modulating inflammatory markers( Reference Meyer, Elmadfa and Herbacek 10 ). In our study, we used a relatively low inoculation of LGG, compared with the work of Kekkonen et al. ( Reference Kekkonen, Lummela and Karjalainen 9 ) in which benefits on fasting inflammatory biomarkers were observed; however, effects on inflammation in healthy populations have been observed at this dosage( Reference Schultz, Linde and Lehn 64 ). Further research on the different bacteria used and their relative dosage could provide insight into the positive effects observed on inflammation in our work and may even reveal benefits on fasting markers of inflammation.
A common reduction after acidified milk and probiotic yogurt intakes was observed for the strain B. wadsworthia. The presence of this strain has been associated with acute infections( Reference Finegold, Summanen and Hunt Gerardo 65 ), and is modulated by probiotic dairy product interventions( Reference Odamaki, Kato and Sugahara 66 ). Veiga et al. ( Reference Veiga, Pons and Agrawal 67 ) recently demonstrated that consumption of a fermented milk product decreased the abundance of this opportunistic pathogen in subjects with irritable bowel syndrome. The pro-inflammatory activity of B. wadsworthia in the presence of saturated fat appears to be mediated by its impact on bile salt metabolism and, consequently, lipid uptake( Reference Devkota and Chang 68 ). Interestingly, B. wadsworthia is increased in mice fed a fat-enriched Western diet( Reference Devkota, Wang and Musch 69 ). Our results using full-fat dairy products are in line with evidence in the scientific community to reappraise the impact of dairy foods and milk fat on CVD risk( Reference German, Gibson and Krauss 70 ).
In our study, metagenomic analysis proved to be a sensitive method that not only showed changes during the interventions but also supported the verification of the compliance of the volunteers with the dairy dietary restrictions. In view of this, we suggest that 16S rRNA PCR-based microbiota analysis could be a useful, accessible tool for the evaluation of compliance in probiotic interventions.
One explanation for the association between a high-fat diet, inflammation and the gut microbiota is the transfer of microbiota-derived LPS during lipid uptake( Reference Cani, Amar and Iglesias 13 , Reference Cani, Possemiers and Van de Wiele 14 ). Whereas we observe a change in bacteria present in the microbiota as well as modulation of inflammatory parameters during our HFM, we were unable to distinguish a significant change in LPS response as persistently low levels of the biomarker were observed on all test days. Given the healthy population studied, it is conceivable that a small change in circulating LPS could lead to larger, measurable changes in circulating interleukins and chemokines. However, these associations may be better elucidated in a population with existing sub-clinical inflammation.
The major strengths of our study are the highly controlled nature of the dietary intervention that included an identical, 3-d controlled diet before each test day. The use of a cross-over design was also an important aspect of the study that facilitated the identification of microbiota changes during the intervention. Given the inter-individual variability in both the baseline microbiota and in the relative changes of the microbiota, use of a parallel study design may not have been adequately sensitive to detect these effects. One unexpected finding in our study was the role of the acidified milk on both the microbiota and the biomarkers of inflammation. This latter effect has not previously been described although there is evidence for the role of other prebiotic foods in modulating inflammation( Reference Lomax and Calder 71 ). Whereas the study design was not specifically adapted to observe an effect of acidified milk, the baseline tests completed during the run-in phase allowed the effects of the acidified milk to be assessed compared with normal milk. In consideration of the anti-inflammatory effects associated with acidified milk, it would be interesting to explore whether the combined use of acidified milk supplemented with probiotic strains could have greater effects of inflammation.
The current study supports a beneficial role of milk supplemented with probiotics and prebiotic ingredients on inflammation associated with a high-fat diet. These benefits may be modulated by modification of the gut microbiota mediated by the probiotic and prebiotic qualities of the tested products. These foods could form a useful part of interventions for populations at risk for inflammation.
Acknowledgements
The authors wish to thank F. Secretan, C. Pellet, Dr W. Kong and Dr M. van Leckwyck for their clinical assistance and support during the clinical test days; R. Blase and co-workers at Agroscope for the microbiological quality control on the dairy products; and, C. Egger and R. Badertscher for their nutritional analyses of the dairy products.
Endotoxin assays were financed by a collaboration with the University of Warwick using a grant received from the Higher Education Innovation Fund 5. The Higher Education Innovation Fund 5 had no role in the design, analysis or writing of this article.
K. J. B., G. P., A. C., G. G., F. P. P., G. V. and N. V. designed the study; K. J. B., M.-J. V. and N. V. conducted the research; G. P., U. B. and U. v. A. produced and assigned the test products; M. R., K. J. B. and U. B. analysed data and performed statistical analyses; K. J. B., M. R., A. C., U. B., G. G., F. P. P., G. V. and N. V. wrote the paper; N. V. and K. J. B. had primary responsibility for the final content; M.-J. V. completed cytokine assays, P. G. M. completed endotoxin assays, J. D. completed NEFA assays, S. A. completed the microbiota preparation and measurement. All authors read and approved the final manuscript.
None of the authors has any conflict of interest to declare.
Supplementary materials
For supplementary material/s referred to in this article, please visit https://doi.org/10.1017/S0007114517000885








