INTRODUCTION
Streptococcus agalactiae, also known as group B streptococcus (GBS) is the leading cause of meningitis, pneumonia and bacterial sepsis in neonates in the USA and Europe [Reference Schrag1]. While this bacterium is known to be carried asymptomatically in the genitourinary and gastrointestinal tracts of up to 35% of healthy adults, it can adversely affect pregnant women, the immunocompromised and the elderly [Reference Farley2, Reference Tettelin3].
β-lactam agents such as penicillin or ampicillin are the antibiotic agents of choice for prophylaxis and treatment of GBS infections. Macrolides are the recommended second-line drugs and the first alternative in cases of β-lactam allergy. Resistance to the alternative drugs of choice for the treatment of GBS infections, including lincosamides such as clindamycin and macrolides such as erythromycin, has increased during the last decade in several countries with some geographical variations [Reference Acikgoz4–Reference Brimil9].
There are two frequently encountered erythromycin resistance mechanisms in streptococci. The first, ribosomal modification is encoded by erm genes which can be inducibly or constitutively expressed, results in cross-resistance to macrolides, lincosamides and streptogramin-B antibiotics [inducible macrolide resistance (iMLSB) and constitutive macrolide resistance (cMLSB) phenotypes, respectively]. Such resistance gives rise to one of two phenotypes, the iMLSB or cMLSB phenotype, respectively. The second mechanism is an active drug efflux pump encoded by a mef gene designated M phenotype (resistance to macrolides only). Seppälä et al. [Reference Seppälä, Nissinen, Quan and Huovinen10] have described a triple disk diffusion test (TDDT) that can be used to characterize GBS isolates into one of these three macrolide resistance phenotypes.
Until recently GBS was classified into nine distinct serotypes based on a capsular polysaccharide antigen, namely Ia, Ib, II, III, IV, V, VI, VII and VIII. In 2007 Slotved et al. proposed a new serotype (ST) designated ST IX [Reference Slotved11]. The predominant serotypes have changed over time, vary with geographical region and ethnic origin and can be associated with different diseases [Reference Brimil9, Reference Davies12–Reference Matsubara17].
This research offers the first insight into the level and types of macrolide resistance in colonizing strains of GBS in Southern Ireland.
METHODS
Bacterial isolates
A total of 324 GBS isolates were collected at the Microbiology Department at Cork University Hospital between 2004 and 2006 by culturing 2000 vaginal swabs from women between the ages of 15 and 54 years on Islam agar (GBS agar base CM755; Oxoid, UK), which was used and interpreted according to the manufacturer's instructions. S. agalactiae MICRO6 and Enterococcus faecalis ATCC 29212 were used as positive and negative culture control strains, respectively. Colonies presumptive for GBS were confirmed by latex agglutination test using the Remel Streptex kit (Launch Diagnostics, UK). All isolates were stored in glycerol on preservative beads at −70°C.
Antimicrobial susceptibility testing
All isolates were tested against erythromycin by disk diffusion using CLSI guidelines for performance and interpretation of susceptibility testing [Reference Wikler18]. The zone sizes around each disk were measured using the BIOMIC Vision Microbiology Analyzer automated callipers (Giles Scientific, USA). Erythromycin minimum inhibitory concentrations (MICs) were determined for the 37 resistant isolates using erythromycin E test according to the manufacturer's instructions (AB Biodisk, Sweden).
Phenotypic characterization
A modified version of the TDDT, as previously described [Reference Seppälä, Nissinen, Quan and Huovinen10], was used to determine the mechanism of erythromycin resistance. Briefly, an erythromycin disk (15 μg) was placed on the centre of the plate with a spiramycin disk (100 μg) 20–25 mm to the left (josamycin was used in the original study) and a clindamycin disk (2 μg) 20–25 mm to the right of it (Oxoid). Plates were incubated for between 20 h and 24 h at 37°C in 5% CO2. Isolates were characterized as possessing the M, iMLSB or cMLSB phenotype using the previously described interpretive criteria [Reference Seppälä, Nissinen, Quan and Huovinen10].
Detection of erythromycin resistance genes
All 37 isolates were investigated for the presence of ermB, ermTR and mefA genes by uniplex PCR. DNA extracts were prepared using the High Pure PCR Template Preparation kit (cat. no. 1796828, Roche Diagnostics, USA) according to the manufacturer's instructions. The primers used and the conditions for amplification of the macrolide resistance genes were as previously described [Reference Sutcliffe19].
The following isolates were used as positive control strains for each PCR: ermB gene Streptococcus pyogenes no. 8902. ermTR gene S. pyogenes no. 8973 and mefA gene S. pyogenes no. 8979. PCR products were resolved on 1·5% agarose gels.
Serotype analysis
All 37 erythromycin-resistant GBS isolates were serotyped using the latex agglutination test according to the manufacturer's instructions. Initially, the Essum Probiotics GBS serotyping kit (Sweden) was used. This kit consists of antisera to serotypes Ia–VIII. Isolates that failed to type using this kit were typed using the Statens Serum Insititut Strep-B latex kit (Denmark). This kit consists of ten GBS antisera, Ia–IX. Isolates that subsequently failed to type using either serotyping kit were deemed non-typable by latex agglutination.
RESULTS
Macrolide resistance rate
Screening of 2000 vaginal swabs yielded a total of 324 GBS isolates – giving a GBS colonization rate of 16·2%. The erythromycin resistance rate was 11·4% (n=37/324). Erythromycin MICs ranged from 2 to >256 μg/ml for the 37 isolates tested (see Table 1).
Table 1. Summary of phenotypic characterization, genetic analysis, serotyping studies and erythromycin minimum inhibitory concentrations (MICs) for the 37 erythromycin-resistant GBS strains isolated from the vaginal flora of women of childbearing age in Southern Ireland
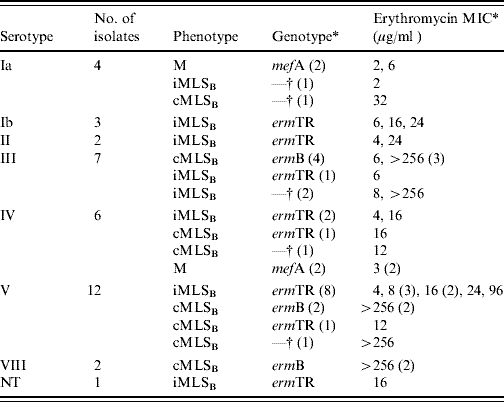
NT, Not typable.
* Values within parentheses represent number.
† Isolates were negative for all three macrolide resistance genes tested namely ermTR, ermB and mefA.
Phenotypic characterization of macrolide resistance
All three macrolide resistance phenotypes were observed as follows: iMLSB (n=20), cMLSB (n=13) and the M phenotype (n=4).
Genotype analysis
The ermTR gene was most frequently expressed (n=19), followed by the ermB gene (n=8). A total of four isolates harboured the mefA gene. Six isolates yielded no PCR products.
GBS serotype distribution
The data obtained on the genotype, phenotype and serotype for each of the 37 erythromycin-resistant GBS isolates collected is given in Table 1. A single isolate was non-typable following latex agglutination.
DISCUSSION
This study yielded a vaginal GBS colonization rate of 16·2% (n=324/2000). These findings are comparable with those seen in other Western European countries, which range between 11% and 21% [Reference Barcaite6].
The erythromycin resistance rate of 11·4% (37/324 isolates) observed in the current study is within the rate ranges reported in Europe (9·2–22·4%) [Reference Acikgoz4, Reference Gherardi7, Reference Portillo8, Reference Aynur20–Reference Figueira-Coelho22]. In Ireland, the implementation of the Royal College of Obstetrics and Gynaecologists' chemoprophylaxis guidelines may in part be responsible for the relatively low level of macrolide resistance seen in this study [23]. These guidelines do not recommend universal antenatal GBS screening and advise intrapartum antibiotic prophylaxis (IAP) only in instances where there has been a previous infant with GBS disease or in cases of GBS bacteruria or GBS colonization in the current pregnancy.
Phenotypic characterization of macrolide resistance has proven a useful tool in determining the antimicrobial susceptibility pattern of an isolate. The predominance of the iMLSB phenotype (54%) found here is in contrast with results reported elsewhere in Western Europe where the cMLSB phenotype is the most prevalent (cMLSB range 52–89% compared to iMLSB range 6–27%) [Reference Portillo8, Reference De Mouy21, Reference Figueira-Coelho22, Reference Diekema24, Reference Culebras25]. However, the iMLSB phenotype is the most common in the USA (48% compared to 27% for cMLSB) and Turkey (80% compared to 20% cMLSB) [Reference Acikgoz4, Reference Heelan, Hasenbein and McAdam26].
The two most studied erythromycin resistance mechanisms are ribosomal modifications and macrolide efflux, encoded by erm and mef genes, respectively. From a clinical perspective the presence of erm genes are a greater cause for concern as the erythromycin ribosomal methylase enzyme can lead to cross-resistance to a wide range of antibiotics due to a mutation at the binding site which alters the affinity of the site for macrolides, lincosamides and streptogramin-B antibiotics. In our study ermTR was the most common macrolide resistance gene in the GBS isolates, followed by ermB. There are very few studies that report a predominance of the ermTR gene in erythromycin-resistant GBS [Reference D'Oliveira27]. However, the low frequency of mefA observed in this research is consistent with data obtained in other countries [Reference Acikgoz4, Reference Gherardi7, Reference Portillo8, Reference Aynur20, Reference Culebras25]. Six isolates yielded no PCR products, phenotypically three were cMLSB and three were iMLSB. Since the discovery of erm and mef genes, macrolide resistance due to mutations (including deletions, insertions and substitutions) of the 23S rRNA and the ribosomal proteins has been observed in other streptococci [Reference Rantala28–Reference Tait-Kamradt30]. These authors propose that macrolide resistance in these six isolates may be attributed to one of these other mechanisms of resistance; however, further study is required for confirmation of this hypothesis.
There was some phenotype–genotype correlation in our study. Genetic analysis showed that the four isolates displaying the M phenotype expressed the mefA gene while 8/13 isolates displaying the cMLSB phenotype expressed the ermB gene. Seventeen of the 20 isolates displaying the iMLSB phenotype expressed the ermTR gene, while two isolates with the cMLSB phenotype expressed the ermTR gene. This has also been observed in research in the USA, where Heelan et al. [Reference Heelan, Hasenbein and McAdam26] found that four isolates displaying the cMLSB phenotype expressed the ermTR gene. These authors hypothesize that ermTR may have mutated so that it can now be constitutively expressed but this requires further investigation.
Of note in this population of isolates is the predominance of the inducible macrolide resistance phenotype. This phenotype cannot be detected using routine antimicrobial susceptibility testing therefore we recommend that the TDDT should be performed on all GBS isolates from penicillin-allergic patients or from patients whose allergy status is unknown.
Typically higher erythromycin MICs are found for cMLSB phenotypes and the results of our study concur with this [62% (8/13), of the cMLSB isolates had MICs >256 μg/ml] [Reference Varman31]. Correlation between phenotype and genotype has been also reported [Reference Poyart32, Reference Von Both33]. Interestingly, in this study 7/8 cMLSB isolates with high MICs harboured the ermB gene, with the last isolate remaining uncharacterized for erythromycin resistance determinant. Lower MICs were found for all M phenotype isolates (range 2–6 μg/ml) and 90% of the iMLSB isolates (2–24 μg/ml) [Reference Varman31].
In the current study serotypes V and III predominated (32% and 19%, respectively), and there is an association between these serotypes and invasiveness [Reference Davies12, Reference Ekelund and Konradsen14, Reference Poyart32]. These results are not unexpected, however, as they are similar to those seen elsewhere [Reference Borchardt5, Reference Figueira-Coelho22, Reference Culebras25].
A surprising finding was that ST IV accounted for 6/37 GBS isolates, a serotype that has not been widely reported globally. ST IV is only commonly seen in colonizing strains in pregnant women in Japan and has infrequently been reported elsewhere [Reference Matsubara17]. Furthermore, the identification of two erythromycin-resistant ST VIII isolates which to date have not been documented elsewhere represents a novel finding in GBS.
Evidence of a serotype–genotype association was observed in this study as shown in Table 1. Serotype–genotype associations have been reported previously. Dogan et al. found that mefA was only expressed in ST Ia isolates, a finding also of our research [Reference Dogan13]. However, ermB was not confined to ST V isolates in this study as has previously been reported [Reference Young34]. A study by Poyart et al. [Reference Poyart32] revealed that ermTR predominated in ST V isolates and these findings were similar to the results observed in the current study.
Erythromycin resistance in GBS has been demonstrated here to be encouragingly low. This study establishes a baseline for monitoring erythromycin resistance in GBS and also provides an insight into serotype distribution in erythromycin-resistant isolates.
CONCLUSION
This study provides an in-depth analysis of the epidemiology of erythromycin-resistant GBS in women of childbearing age in Southern Ireland. Erythromycin resistance in GBS has been demonstrated here to be encouragingly low (11·4%). This investigation provides data which establishes a baseline for monitoring erythromycin resistance in GBS in this region in the future and also provides an insight into serotype distribution in these erythromycin-resistant isolates. The serotype analysis herein reveals that whilst the most prevalent serotypes are similar to those seen elsewhere (ST III and ST V) that other less common serotypes (such as ST IV) are also present. As our societies become more culturally diverse, continued surveillance of GBS serotype distribution is necessary to aid in the development of an effective GBS serotype-based vaccine.
ACKNOWLEDGEMENTS
We acknowledge the Department of Medical Microbiology, Cork University Hospital for the provision of anonymized samples and consumables. Thanks are also due to J. Melo-Cristino (Institute of Microbiology, Lisbon School of Medicine, Lisbon, Portugal) for provision of control strains. We acknowledge the financial support from Cork Institute of Technology ‘seed fund’ for completion of this project. In addition financial support received by R. Kiely as fees from the Health Service Executive is gratefully acknowledged.
DECLARATION OF INTEREST
None.



