INTRODUCTION
Escherichia coli, a commensal of the intestinal tract of humans, as well as many animals, is a Gram-negative, facultative anaerobic rod. The virulent, disease-causing E. coli can be divided into two major groups: gastrointestinal and extragastrointesinal pathogens. The gastrointestinal category includes enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), enteroinvasive E. coli (EIEC), enteroaggregative E. coli (EAEC) and Shiga toxin-producing E. coli (STEC), which are significant causes of foodborne diseases. STEC can lead to severe illness, like haemolytic uraemic syndrome (HUS), which presents with haemolytic anaemia, thrombocytopenia and renal insufficiency, most often after an onset of diarrhoea (bloody or non-bloody). In many different O:H serotypes, there is a group of the so-called top five serogoups (O26, O103, O111, O145, O157) which are mainly related to severe illness in humans. Although the importance of non-O157 serovars is being increasingly recognized, so far O157 has caused most outbreaks and cases of severe disease throughout the world [Reference Hedican1–Reference Blanco3].
A stepwise evolutionary model postulates that the O157 clonal group splits into one lineage, leading to the common non-sorbitol-fermenting (nSF) O157:H7 clonal complex, and a second branch of sorbitol-fermenting (SF) O157:H− (non-motile) strains [Reference Feng4]. Recently, SF STEC O157:H− strains have been increasingly isolated from clinical cases [Reference Bielaszewska5, Reference Friedrich6], emphasizing the importance of analysing patients' stool samples for both SF and nSF stx-producing E. coli O157.
E. coli O157:H7/H− exist in humans and in the environment as stx-positive and stx-negative variants [Reference Friedrich6, Reference Bielaszewska7]. Friedrich et al. [Reference Friedrich6] compared the prevalence of stx-negative strains in SF and nSF E. coli O157 isolated from stools of patients with sporadic cases of diarrhoea or HUS. They found that the proportion of stx-negative strains was significantly higher in SF O157:H− isolates (12·7%) than in nSF O157:H7/H− isolates (0·8%). These authors also identified stx-negative SF E. coli O157 strains in patients' stool samples during three outbreaks of HUS and one outbreak of diarrhoea and documented loss of stx genes during outbreaks of human infection [Reference Friedrich6].
Since few specific data of clinical human O157 STEC strains are available in Switzerland, the aim of this study was (i) to further characterize all O157 STEC strains sent to the National Centre for Enteropathogenic Bacteria (NENT) from 2000 to 2009, and (ii) to compare the results with data from other countries.
METHODS
Strains
A total of 144 STEC strains collected and stored (−80°C) by NENT were used for further characterization. The strains were isolated between 2000 and 2009 from human faecal samples, collected by hospitals or family doctors. All strains were confirmed to be STEC by detection of stx genes by polymerase chain reaction (PCR) with primers VT1 and VT2, based on sequence targeting of a conserved region between stx1 and stx2 genes and PCR conditions described previously [Reference Zweifel and Stephan8].
Serotyping
O157 strains were distinguished from the collection of the 144 STEC strains using PCR with primers based on sequences of the rfbE (per) gene [Reference Abdulmawjood, Roth and Bulte9]. STEC O157:H7 strain EDL 933 was included as a control. The H antigen of the O157 strains was determined by PCR with primers FLICH7-F and FLICH7-R, and PCR conditions as described previously [Reference Gannon10]. Moreover, all O157 strains were serotyped with an O157:H7 latex agglutination test (Wellcolex E. coli O157:H7; Remel, USA).
Further strain characterization
Fermentation of sorbitol was detected on sorbitol MacConkey agar (SMAC) (Oxoid Ltd, UK). Strains were further tested by PCR for stx1 and stx2 [Reference Russmann11], eae and eae γ1 encoding intimin [Reference Blanco12], and ehxA encoding EHEC haemolysin [Reference Schmidt, Beutin and Karch13]. Further characterization of the Shiga toxin type 2 variant B-subunit was done by PCR–RFLP [Reference Stephan and Hoelzle14].
Phage-typing
Bacterial phage-typing was performed at the Laboratory of Gastrointestinal Pathogens, GEZI (HPA Centre for Infections, London, UK) by the methods described by Khakhria et al. [Reference Khakhria, Duck and Lior15].
Multi-locus sequence typing (MLST)
Internal amplicons of seven housekeeping genes (adk, fumC, gyrB, icdF, mdh, purA, recA) were sequenced [Reference Wirth16] and alleles as well as sequence types (ST) were assigned in accordance with the E. coli MLST website (http://mlst.ucc.ie/mlst/dbs/Ecoli).
Genotyping
Pulsed-field gel electrophoresis (PFGE) was performed according to the CDC PulseNet protocol (http://www.cdc.gov/pulsenet/protocols.htm) with minor modifications. Briefly, strains were grown on blood agar at 37°C overnight. Colonies from blood agar were resuspended in cell suspension buffer (OD600=1). The bacterial cell suspension was mixed with 400 μl of 1·4% Bio-Rad agarose (Bio-Rad, Germany) and cells were lysed by proteinase K treatment overnight. After lysis the plugs were washed twice for 15 min in ultrapure water and four times for an hour in Tris-EDTA (TE) buffer. After washing with TE buffer, DNA agarose plugs were incubated overnight in the presence of XbaI (Roche, Germany) according to the manufacturer's instructions. Restricted DNA in plug slices was separated in 1% SeaKem Gold (BioConcept, Switzerland) agarose gel at 6 V/cm in 0·5× Tris-borate-EDTA buffer cooled to 14°C in a CHEF-DR III system (Bio-Rad). The pulse times were ramped from 5 s to 50 s for 20 h at an angle of 120°. Gels were stained with ethidium bromide and visualized under UV light transillumination with Gel Doc (Bio-Rad) and analysed with BioNumerics software (Applied Maths, Belgium).
As a reference Salmonella Braenderup strain H9812 (ATCC BAA 664) was used.
Antimicrobial susceptibility testing
The strains were tested for antimicrobial resistance by the disk diffusion method according to the protocols of the Clinical Laboratory Standards Institute (CLSI, 2008). The panel of antibiotics (disks: Becton, Dickinson, USA) consisted of ampicillin (AM), amoxicillin/clavulanic acid (AMC), ceftazidime (CAZ), cefalothine (CF), ciprofloxacin (CIP), cefpodoxime (CPD), cefotaxime (CTX), cefuroxime (CXM), cefepime (FEP), cefoxitin (FOX), gentamicin (GM) and tetracycline (Te). E. coli ATCC 25 922 was used as a quality control strain. The strains were classified as resistant or susceptible to each antibiotic agent. Strains giving ‘intermediate’ values were considered susceptible.
Results
Medical history data
Of the 144 STEC strains collected during 2000–2009, 44 strains from 44 different patients were identified as O157. Medical history data for these patients are summarized in Figure 1. Thirteen patients (29·5%) developed HUS, of which six (46·2%) were male, and seven (53·8%) female, with an average age of 3·5 years (range 1–15 years). Twenty-seven (61·4%) patients presented with bloody diarrhoea, seven (15·9%) with non-bloody diarrhoea, and four (9·1%) were anaemic. For nine patients no medical history data was available. Thirty-three (76·7%) patients were aged ⩽10 years, three (7%) were aged ⩾60 years and seven (16·3%) patients were aged between 10 and 60 years. For one patient (female, presenting with HUS, diarrhoea and anaemia) no data was available.
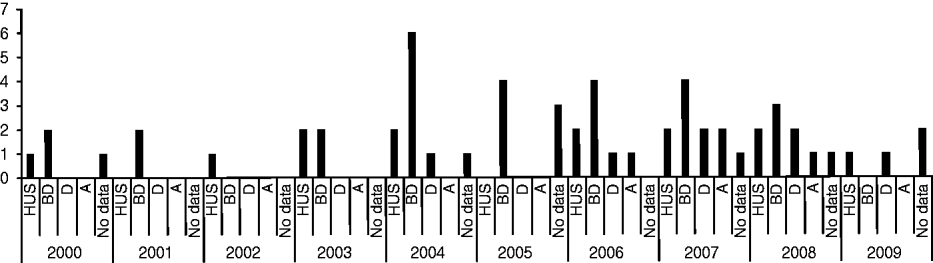
Fig. 1. Medical history data of 44 STEC O157 strains isolated from human patients from 2000 to 2009 in Switzerland. HUS, Haemolytic uraemic syndrome; BD, bloody diarrhoea; D, non-bloody diarrhoea; A, anaemia.
Further strain characterization
Latex agglutination of all 44 strains was positive for O157, whereas 11 (25%) strains were negative for H7 agglutination (O157:NM). Results are listed in Table 1. Nevertheless, these strains were positive for the fliC gene by PCR.
Table 1. Serotypes and virulence factors of STEC O157 strains isolated from human patients from 2000 to 2009 in Switzerland

[H7], Negative by latex agglutination serotyping, positive by PCR for fliC; PT, phage type; RDNC, Reacts but Does Not Confirm to a published typing pattern; ST, sequence type; Te, tetracycline; A, anaemia; BD, bloody diarrhoea; D, non-bloody diarrhoea; HUS, haemolytic uraemic syndrome.
Four (9·1%) strains fermented sorbitol (SF) on SMAC, 40 (90·9%) were nSF. All four SF strains were O157:H−, 33 (82·5%) of the nSF strains were O157:H7, and seven (17·5%) strains were O157:H−. All 44 E. coli O157 strains tested positive for eae and ehxA genes. Among the strains, 52·3%, 63·6%, 15·9%, and 20·5% harboured stx1, stx2, stx2c, and the combination of stx2 and stx2c, respectively (Table 1).
Phage-typing
Nine different phage types were found (PT2, 4, 8, 14, 23, 32, 49, 50, 71), and six (13·6%) strains could not be further characterized by phage-typing (RDNC, Reacts but Does Not Conform to a published typing pattern). PT2, 4, 14, 50 and 71 appeared only once. PT8 was found eight times (18·2%), PT23 twice (4·5%), PT32 nineteen times (43·2%) and PT49 resulted in four (9·1%) of the strains (Table 1).
MLST
All 44 strains belonged to ML ST 11, similar to the vast majority of eae-positive human STEC O157 isolates.
PFGE typing
PFGE patterns were very heterogeneous except for eight strains (18·2%). The remaining 36 patterns showed similarity coefficients between 59% and 94% (Dice similarity index and UPGMA method). The dendrogram is displayed in Figure 2.

Fig. 2. PFGE dendrogram (Dice similarity index and UPGMA method) of 44 STEC O157 strains isolated from human patients from 2000 to 2009 in Switzerland. PT, Phage type; RDNC, Reacts but Does Not Confirm to a published typing pattern; stx2, stx2 variants (stx2 and/or stx2c).
Antimicrobial susceptibility testing
All O157 STEC strains were susceptible to 11 or more antibiotic agents (AM, AMC, CAZ, CF, CIP, CPD, CTX, CXM, FEP, FOX, GM). Of the 44 strains, two (4·5%) were resistant to Te.
Discussion
The 44 strains investigated in this study are highly representative for Switzerland. This is the total number of STEC O157 pathogens isolated by NENT from all human material submitted over the 10-year period (2000–2009). Switzerland is a small country in the centre of Europe that is multi-cultural and thus suffers epidemiological influences from several neighbouring ethnic backgrounds. Moreover, the Swiss are known to be the most travelled people in the world. With a population of only 7·1 million, Swiss people execute about 2·2 million trips abroad per year. These aspects also warrant international relevance for our data.
HUS, as a consequence of STEC O157 infection, was recorded in 13 (29·5%) of the 44 investigated patients. Of patients with HUS 84·6% were aged ⩽5 years. Of the HUS patients, six (46·2%) were male, seven (53·8%) were female. A similar age and gender distribution was reported in a study spanning a 4-year period in Germany [Reference Ammon, Petersen and Karch17].
In contrast, in a study performed over a 7-year period (2000-2006) involving 3464 STEC O157 isolates in Northern America, the authors observed fewer HUS cases (6·3%) and fewer HUS patients aged <5 years (15·3%) [Reference Gould18]. Similar data to these were obtained in Minnesota, Wales and England [Reference Hedican1, Reference Chalmers19, Reference Willshaw20].
Of the 44 STEC O157 strains, 47·7% harboured stx2 variants only and 52·3% possessed a combination of stx1 and stx2 variant genes. No strain was positive for stx1 only. A similar distribution of stx genes was found for clinical STEC O157 isolates in different countries [Reference Hedican1, Reference Blanco3, Reference Chalmers19–Reference Mora22]. In our study, in all HUS cases strains harbouring the stx2 gene type were involved. Nevertheless, there was no evidence that the stx2 variants or the combination of stx1, stx2 and stx2c genes are directly associated with the clinical outcome.
All 44 STEC O157 strains were positive for ehxA and eae irrespective of the clinical symptoms. Orth & Wurzner [Reference Orth and Wurzner21] gained comparable results with all isolates investigated in their study being positive for stx2, eae and ehxA. Similar to other studies [Reference Blanco3, Reference Beutin23], intimin gamma was the eae subtype in our O157:H7 strains.
For years STEC O157 has been classically mentioned as being nSF and motile. In 1990 an outbreak of HUS in Germany, for the first time yielded SF O157:H− [Reference Karch24]. This loss of motility in SF STEC O157 is caused by a 12-bp in-frame deletion in fliC that is required for transcriptional activation of genes involved in flagellum biosynthesis [Reference Monday, Minnich and Feng25]. Although a low prevalence of SF O157 strains were found in our strain collection, their occurrence warrants the use of additional methods other than SMAC.
In Germany SF STEC O157:H− are the second most common cause of sporadic HUS [Reference Karch, Tarr and Bielaszewska26] and have, up to 2005, caused two large outbreaks involving 57 HUS cases of which seven children died [Reference Karch, Tarr and Bielaszewska26].
Among our isolates four (9·1%) SF strains and seven (15·9%) nSF strains were non-motile. HUS developed from 11 classical nSF O157:H7, one atypical SF O157:H− and one nSF O157:H−. Of the other three patients with SF STEC O157 isolates, two developed bloody diarrhoea, and from one, no data was available. Patients with SF STEC O157 were aged 1, 2, 5 and 7 years. In Germany, SF STEC O157 was present in 13·3–40·5% of HUS patients, in 7·4–25% of patients with diarrhoea and only in children aged <3 years [Reference Karch and Bielaszewska27]. All SF STEC O157:H− strains harboured the fliC gene-encoding H7 antigen, as in our study.
To further characterize our strains we performed phage-typing and PFGE. Among our STEC O157 isolates nine different phage types were found, with PT8, PT32 and PT49 the most prevalent. In a study on 415 STEC O157 patients in Wales, the most common among 19 detected phage types were PT2 and PT49 showing frequencies of 42·3% and 12·7%, respectively [Reference Chalmers19]. Similar results were found in an English study [Reference Thomas28], where PT2 (46%) was the most common phage type, followed by PT49 (17%) and PT8 (8%). In a recent study from Spain, PT2 was also the most frequently found phage type in human STEC O157, followed by PT8 [Reference Mora22]. Therefore, it appears that Switzerland has a different situation.
PFGE patterns were very heterogeneous except for eight strains (18·2%). Of these eight strains there were two sets each consisting of two strains with similar patterns (set 1: strain K 02-04, male, aged 1 year, and strain K 774-04, male aged 5 years, both strains isolated in 2004, both stx2, eae positive and PT23; set 2: strain K 1787-02, male aged 2 years, sample isolated in 2002, and strain K 1206-06, female aged 45 years, sample isolated in 2006, both stx1, stx2, eae positive and PT32) and one set consisting of four equal patterns, all four PT32 and stx1, stx2 and eae positive (K 447-04, female aged 10 years, sample isolated in 2004 and K 1097-05, male aged 2 years, K 1127-05, male aged 7 years and K 1144-05, female aged 40 years, all three samples isolated in 2005). The high genetic diversity within the strains leads to the conclusion that STEC O157 infections in Switzerland most often occur as single cases. Similar to the situation in Switzerland, only a few STEC O157:H7-associated outbreaks have been reported in the neighbouring countries within this time period [Reference Alpers29–Reference King31].
In our study a very low prevalence of antibiotic resistance was found for STEC O157 isolates. Only two strains (4·5%) were resistant to tetracycline. In England and Wales 20% of human STEC O157 strains collected between 1995 and 1998 were resistant to one or more antimicrobial agents [Reference Willshaw20]. Only 1% of human STEC showed resistance in Germany [Reference Schmidt32]. Recently, Srinivasan et al. [Reference Srinivasan33] investigated 153 STEC O157:H7/H− strains isolated from human faeces, cows and food in the USA of which >90% showed resistance to ampicillin and cephalothin. Moreover, resistance to tetracycline, gentamicin, cefotaxime and ciprofloxacin was found. High prevalence of resistance was also found in O157:H7 strains from Spanish people [Reference Mora22], where 38% and 24% of the isolates showed resistance to tetracycline and ampicillin, respectively.
To summarize, only 30·6% of the STEC strains isolated from clinical cases from 2000 to 2009 in Switzerland were STEC O157. SF and nSF strains were found. All strains were positive for stx2 variants (stx2 and/or stx2c), eae and ehxA and showed a very favourable antibiotic resistance situation. The high genetic diversity within the strains leads to the conclusion that STEC O157 infections in Switzerland most often occur as sporadic cases.
ACKNOWLEDGEMENTS
We thank the FOPH (Federal Office of Public Health) for support in collecting the medical history data and Grethe Sägesser for help in strain collection and technical support.
DECLARATION OF INTEREST
None.





