1. Introduction
Bone Morphogenetic Protein (BMP) signalling ligands regulate numerous cellular processes in multi-cellular animals, including growth, differentiation, migration and death (Chen et al., Reference Chen, Zhao and Mundy2004). In the regulation of growth, these ligands may act at the level of tissue, organ and/or organism size. For example, in Drosophila, the BMP-related ligand Dpp is also involved in growth regulation. In the wing imaginal disc, reduction of Dpp activity results in reduced wing size and clones of cells deficient in Dpp signalling components fail to proliferate (Burke & Basler, Reference Burke and Basler1996; Lecuit et al., Reference Lecuit, Brook, Ng, Calleja, Sun and Cohen1996). Conversely, elevated Dpp pathway activation can lead to a striking overgrowth phenotype (Burke & Basler, Reference Burke and Basler1996; Lecuit et al., Reference Lecuit, Brook, Ng, Calleja, Sun and Cohen1996; Nellen et al., Reference Nellen, Burke, Struhl and Basler1996). In promoting the growth of the imaginal disc, Dpp acts by regulating cell proliferation rather than cell size. Similarly, vertebrate BMPs were first identified by their ability to stimulate bone morphogenesis (Wozney et al., Reference Wozney, Rosen, Celeste, Mitsock, Whitters, Kriz, Hewick and Wang1988), and mutations in vertebrate BMPs and their signalling components lead to disrupted skeletal development. For example, mutations in Growth and Differentiation Factor 5 (GDF5) or in the BMP type I receptor, BMPR1B/Alk6, are associated with chondrodysplasia of the limbs and limb joints, and with brachydactyly of the fingers and toes (Thomas et al., Reference Thomas, Lin, Nandedkar, Camargo, Cervenka and Luyten1996; Everman et al., Reference Everman, Bartels, Yang, Yanamandra, Goodman, Mendoza-Londono, Savarirayan, White, Graham, Gale, Svarch, Newman, Kleckers, Francomano, Govindaiah, Singh, Morrison, Thomas and Warman2002; Lehmann et al., Reference Lehmann, Seemann, Stricker, Sammar, Meyer, Suring, Majewski, Tinschert, Grzeschik, Muller, Knaus, Nurnberg and Mundlos2003). In addition, mutations in the BMP type I receptor ACVR1 (Alk4) result in fibrodysplasia ossificans progressiva, a genetic disorder characterized by ectopic osteogenesis (Shore et al., Reference Shore, Xu, Feldman, Fenstermacher, Brown and Kaplan2006). The mutation is proposed to result in a constitutively active receptor that promotes excessive skeletogenesis. Finally, allelic variants in the genes for BMPs and their negative regulators contribute to differences in height in human populations (Gudbjartsson et al., Reference Gudbjartsson, Walters, Thorleifsson, Stefansson, Halldorsson, Zusmanovich, Sulem, Thorlacius, Gylfason, Steinberg, Helgadottir, Ingason, Steinthorsdottir, Olafsdottir, Olafsdottir, Jonsson, Borch-Johnsen, Hansen, Andersen, Jorgensen, Pedersen, Aben, Witjes, Swinkels, den Heijer, Franke, Verbeek, Becker, Yanek, Becker, Tryggvadottir, Rafnar, Gulcher, Kiemeney, Kong, Thorsteinsdottir and Stefansson2008; Lettre et al., Reference Lettre, Jackson, Gieger, Schumacher, Berndt, Sanna, Eyheramendy, Voight, Butler, Guiducci, Illig, Hackett, Heid, Jacobs, Lyssenko, Uda, Boehnke, Chanock, Groop, Hu, Isomaa, Kraft, Peltonen, Salomaa, Schlessinger, Hunter, Hayes, Abecasis, Wichmann, Mohlke and Hirschhorn2008; Weedon et al., Reference Weedon, Lango, Lindgren, Wallace, Evans, Mangino, Freathy, Perry, Stevens, Hall, Samani, Shields, Prokopenko, Farrall, Dominiczak, Johnson, Bergmann, Beckmann, Vollenweider, Waterworth, Mooser, Palmer, Morris, Ouwehand, Zhao, Li, Loos, Barroso, Deloukas, Sandhu, Wheeler, Soranzo, Inouye, Wareham, Caulfield, Munroe, Hattersley, McCarthy and Frayling2008). In contrast to these growth promoting ligands, the BMP-related ligand myostatin is a negative regulator of skeletal muscle mass, inhibiting both cell proliferation and cell growth (McPherron et al., Reference McPherron, Lawler and Lee1997). Belgian Blue cattle, bred for their increased muscle mass, carry mutations in the myostatin gene (McPherron & Lee, Reference McPherron and Lee1997). Thus, in different contexts, BMP ligands can control tissue, organ and organismal growth by regulation of cell proliferation, cell growth, or both.
In the nematode Caenorhabditis elegans, a major growth regulating factor is the BMP signalling ligand DBL-1 (Dpp- and BMP-like-1) (Suzuki et al., Reference Suzuki, Yandell, Roy, Krishna, Savage-Dunn, Ross, Padgett and Wood1999; Savage-Dunn et al., Reference Savage-Dunn, Maduzia, Zimmerman, Roberts, Cohen, Tokarz and Padgett2003). In loss of function mutants for dbl-1 or for any of its signalling components, postembryonic and adult growth are reduced, resulting in small body size (Savage-Dunn et al., Reference Savage-Dunn, Tokarz, Wang, Cohen, Giannikas and Padgett2000). This defect in body size is not due to reduced cell number, but due to reduced cell size. To understand how DBL-1 regulates growth, it is critical to identify the cellular source(s) of the ligand as well as the relevant target tissue(s). We have previously analysed the cellular targets of DBL-1 by determining the spatial requirements for expression of the signalling component Small body size-3 (SMA-3) Smad. The sma-3 gene is expressed primarily in three tissues: the pharynx, the intestine and the hypodermis. By specifically expressing sma-3 in each of these tissues in a sma-3 mutant background, we determined that expression in the hypodermis is necessary and sufficient for normal body size (Wang et al., Reference Wang, Tokarz and Savage-Dunn2002). Consistent with this conclusion, the expression of DBL-1 receptors has also been found to be required in the hypodermis (Inoue & Thomas, Reference Inoue and Thomas2000; Yoshida et al., Reference Yoshida, Morita, Mochii and Ueno2001). The hypodermis is the epidermal tissue that surrounds the nematode body and secretes the cuticle. Most of the hypodermis consists of a large multinucleate syncytium, hyp7.
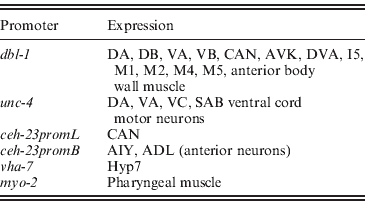
Having identified the epidermis as the critical target tissue for DBL-1 in growth regulation, we now ask where DBL-1 must be expressed to regulate body size appropriately. The dbl-1 gene is expressed primarily in neurons, including ventral cord neurons DA, DB, VA and VB; the lateral excretory canal-associated CAN cells; and anterior neurons AVK, DVA, I5, M1, M2, M4 and M5 (Suzuki et al., Reference Suzuki, Yandell, Roy, Krishna, Savage-Dunn, Ross, Padgett and Wood1999). In addition, dbl-1 expression is seen in anterior body wall muscle. It has not yet been determined, however, whether dbl-1 expression in these identified cells is necessary and/or sufficient for body size regulation. To answer this question, we have directed expression of dbl-1 in particular cells in a dbl-1 mutant background using characterized cell-specific promoters.
2. Materials and methods
(i) Expression constructs
Promoter fragments were cloned upstream of a PstI-BamHI genomic fragment containing dbl-1 coding regions in pBLUESCRIPT SK. The following promoter fragments were generated by PCR (primer sequences available upon request): dbl-1, 4·5 kb SalI-PstI; unc-4, 2·9 kb KpnI-EcoRV; ceh-23promL (CAN), 500 bp HindIII-PstI; ceh-23promB, 2·4 kb EcoRI. The vha-7 and myo-2 promoters were as previously described (Wang et al., Reference Wang, Tokarz and Savage-Dunn2002).
(ii) Transgenic animals
For each construct, 10 ng/μl of plasmid DNA was microinjected into the gonadal syncytia of dbl-1(wk70) mutant hermaphrodites with 100 ng/μl of pRF4 (containing rol-6) as a marker (Mello et al., Reference Mello, Kramer, Stinchcomb and Ambros1991). Transgenic animals were identified by the dominant roller phenotype. For each construct, one representative strain was chosen to quantify.
(iii) Body length measurements
Embryos were harvested from gravid hermaphrodites by bleaching. These were seeded onto plates and grown at 20°C for 96 h until adulthood. Transgenic animals from each strain (n=29–35) were mounted onto slides and photographed using Axio Vision 3.00 software. The length of each worm was determined by drawing a segmented line along the midline using the same software. Statistical significance was determined by Student's t test.
(iv) Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
Since adult body size would be the cumulative effect of dbl-1 expression throughout development, dbl-1 expression levels were determined in mixed stage populations. Animals were collected and total RNA extracted by Trizol (Invitrogen) as described (Liang et al., Reference Liang, Lints, Foehr, Tokarz, Yu, Emmons, Liu and Savage-Dunn2003). SuperScript™ III platinum two-step qRT–PCR kit with SYBR Green (Invitrogen) was used to perform RT–PCR. Real time RT–PCR was performed on a LightCycler 2.0 (Roche). Data analysis software used was LightCycler software version 4.0. Actin gene act-1 was used as standard control. A primer in an intron of the act-1 gene was used to confirm the absence of genomic DNA in RNA preps. Primer sequences available upon request.
3. Results
(i) Transgenic expression of dbl-1 can substitute for endogenous expression to regulate body size
A genomic clone containing all of the dbl-1 coding sequences but lacking upstream sequences was constructed. When this clone was introduced into dbl-1 mutant animals by microinjection, it failed to rescue the small body size phenotype (Figs 1 a and 2), as expected since this clone does not contain the upstream regulatory sequences required for expression. As a positive control, we next constructed a plasmid in which the native dbl-1 promoter is driving expression of dbl-1 coding sequences. When introduced into dbl-1 mutant animals, this construct provides adequate wild-type dbl-1 activity to rescue the body size phenotype (Figs 1 b and 2).

Fig. 1. Adult transgenic animals carrying dbl-1 expression constructs in an otherwise dbl-1(−) mutant background. (a) No-promoter control. (b) dbl-1 native promoter positive control. (c) ceh-23 AIY/ADL neuron promoter element B. (d) unc-4 ventral cord neuron promoter. (e) ceh-23 CAN promoter element L. (f) vha-7 hypodermal promoter. (g) myo-2 pharyngeal muscle promoter. All images were taken at the same magnification (100×).
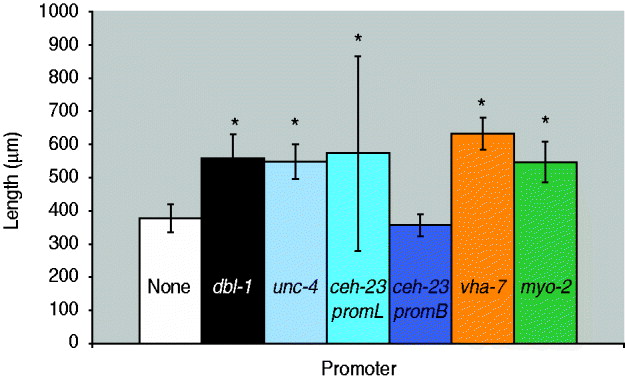
Fig. 2. Quantification of body length of dbl-1 transgenic animals. White bar: no-promoter control; black bar: dbl-1 promoter control; blue bars: neuronal promoters unc-4, ceh-23promL (CAN), ceh-23promB; orange bar: hypodermal promoter vha-7; green bar: pharyngeal muscle promoter myo-2. Error bars show standard deviation. *Significantly different from no-promoter control (P<0·001).
(ii) Non-overlapping subsets of the endogenous dbl-1 expression pattern are sufficient for body size rescue
We next asked whether expression in a subset of the normal dbl-1 expression pattern is sufficient to promote normal body size. We chose three subsets of dbl-1-expressing cells to test: ventral cord neurons, anterior neurons and lateral excretory canal-associated CAN cells. Expression in these subsets of neurons was driven by the unc-4 promoter (Miller & Niemeyer, Reference Miller and Niemeyer1995), the ceh-23 promoter element B and the ceh-23 promoter element L (Wenick & Hobert, Reference Wenick and Hobert2004), respectively (Table 1). We found that dbl-1 expression driven by the unc-4 promoter provides rescue of body size (Figs 1 d and 2) that is indistinguishable from that of the native dbl-1 promoter (P=0·47). Thus, expression of dbl-1 in the ventral cord is sufficient for body size regulation in the absence of expression from the other endogenous sites. Similarly, the expression of dbl-1 in the two CAN cells, in the absence of endogenous expression, provided significant rescue relative to the no-promoter control (P=0·0005) that is not significantly different from that of the dbl-1 native promoter (P=0·79). Thus, dbl-1 expression in the CAN cells, similar to expression in the ventral cord, is also sufficient to support growth to normal size of the animal. In this case, however, a highly variable body size phenotype occurs with large deviations in both the length and the width of animals (Fig. 1 e). The variable body size phenotype was seen in multiple independent lines for this construct (data not shown). This variability in body size is reflected in an unusually broad standard deviation for the quantified body lengths (Fig. 2). Finally, in contrast to the ventral cord and CAN cells, expression from the ceh-23 promoter element B, which drives expression in a small number of anterior neurons, was insufficient to rescue body size (Figs 1 c and 2).
Table 1. Sites of expression of characterized tissue-specific promoters in C. elegans
(iii) Autocrine DBL-1 signalling is efficient as a stimulator of nematode growth
We next tested whether ectopic expression of dbl-1 could replace endogenous expression. We and others have previously shown that the target tissue for DBL-1 activity is the large epidermal syncytium hyp7. Although dbl-1 is not normally produced by hyp7, we used the vha-7 promoter (Wang et al., Reference Wang, Tokarz and Savage-Dunn2002) to drive its expression in hyp7 ectopically and found that this site of expression provides sufficient DBL-1 activity to significantly rescue body size defects of dbl-1 mutants (Figs 1 f and 2). In fact, the expression of dbl-1 in the epidermis allowed significantly better rescue of body size than did expression from the native dbl-1 promoter (P=3·6×10−6). We draw two inferences from this result. First, DBL-1 can be synthesized and secreted in an active form when expressed in epidermal tissue. This precludes the existence of specialized modifying or trafficking mechanisms either in the specific cells that normally express DBL-1 or in neuronal and muscle cells in general. Second, autocrine signalling (epidermis to epidermis) is sufficient to regulate body size and may even be more efficient than the paracrine signalling that occurs under endogenous conditions.
(iv) Localized expression of dbl-1 can stimulate normal body size regulation
Thus far, our results suggest that few constraints exist on the site(s) of expression of dbl-1. The only site of expression that gave a complete lack of rescue was the expression in two pairs of anterior neurons driven by the ceh-23 promoter element B. We therefore considered the possibility that this site of expression failed to produce rescue of body size because DBL-1 may not act at long range, since all of the other expression constructs tested drive expression along the anterior–posterior axis. To express dbl-1 in another anterior-specific domain, we used the myo-2 promoter (Okkema et al., Reference Okkema, Harrison, Plunger, Aryana and Fire1993) that expresses only in the muscle of the pharynx, an anteriorly located organ. In contrast to the result with ceh-23 promoter B, expression from the myo-2 promoter was sufficient to rescue the body size of dbl-1 mutants (Figs 1 g and 2) to a degree that is indistinguishable from that of the native promoter (P=0·52). We therefore conclude that expression of dbl-1 from a localized anterior source does not prevent its regulation of body morphology throughout the length of the animal, indicating that DBL-1 activity is not limited to a short-range action. It is possible that DBL-1 is capable of diffusing along the length of the animal to coordinate growth. Alternatively, since the target epidermal tissue hyp7 consists of a single large multinucleate syncytium, it is possible that activated signal transducers such as Smads may spread efficiently throughout the syncytial tissue even if the ligand remains localized at one end of the anterior–posterior axis.
(v) Expression level, rather than site of expression, may be the critical factor in body size regulation by DBL-1
We have seen that expression of dbl-1 from multiple tissues is sufficient to rescue body size. We next considered the hypothesis that rescue, rather than being dependent on the source of DBL-1, is instead proportional to the expression level. There is precedent for this hypothesis, since DBL-1 is a dose-dependent regulator of growth: dbl-1 loss of function causes small body size, its overexpression results in long body size and heterozygous mutants have intermediate phenotypes (Suzuki et al., Reference Suzuki, Yandell, Roy, Krishna, Savage-Dunn, Ross, Padgett and Wood1999). To test this hypothesis in the context of dbl-1 heterologous expression, we chose two transgenic lines with different rescue phenotypes, the CAN-expressing and hyp7-expressing lines, and integrated the transgenes by γ-irradiation. We then quantified the dbl-1 expression levels by qRT–PCR and compared with the respective body size phenotypes. We first noted that the integration of the transgene eliminated the extreme variability in body size caused by the CAN-expressing extrachromosomal array. Since this transgene directs expression in only two cells, it may have been particularly sensitive to mosaicism due to loss of the array. Quantification of the dbl-1 transcript in these strains demonstrates that expression from the vha-7 promoter in hyp7 was 4-fold higher than the level reached by the CAN-specific promoter (Fig. 3). Furthermore, the strain expressing DBL-1 in hyp7 was significantly longer than the one expressing DBL-1 solely in the CAN cells (Fig. 3). This result is consistent with the interpretation that DBL-1 expression levels determine the level of body size rescue.
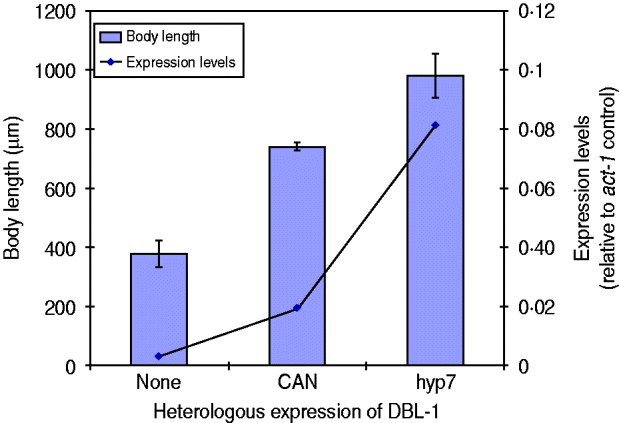
Fig. 3. DBL-1 expression levels may drive body size phenotypes. Blue columns show body length; error bars show standard deviation. Expression of dbl-1 transcript, measured by qRT–PCR, is given in comparison with the act-1 gene control.
4. Discussion
In summary, we have tested the requirements for dbl-1 expression in the regulation of body size in C. elegans. We find that no specific site of expression is necessary for DBL-1 function, and that several non-overlapping sites of expression are sufficient. These sites include endogenous sites of dbl-1 expression (ventral cord neurons and CAN cells) as well as ectopic sites (hyp7 epidermis and pharyngeal muscle). Finally, although the normal expression pattern of dbl-1 is well distributed along the anterior/posterior axis, anteriorly localized expression in the pharynx is sufficient for normal regulation of body size. Given these relaxed requirements for dbl-1 expression, one may consider whether any advantages derive from the endogenous expression pattern. One factor that likely contributes to framing the endogenous expression pattern is the need for dbl-1 to regulate other aspects of C. elegans development and homoeostasis, such as innate immunity and male tail patterning (Savage-Dunn, Reference Savage-Dunn2005; Zugasti & Ewbank, Reference Zugasti and Ewbank2009). A second factor may be that the endogenous pattern provides more robust, consistent control of body size than would be possible if dbl-1 were expressed, for example, from only a subset of neurons such as the CAN cells. Finally, although we found that autocrine signalling from the epidermis promoted increased growth relative to paracrine signalling under laboratory conditions, it is possible that the native expression pattern provides more points of regulation for different environmental conditions and this may provide a fitness advantage under some circumstances.
We are grateful to Dr Iva Greenwald for use of a caesium source. This research was supported by RSG-98-230-04-DDC from the American Cancer Society and by NIH 1R15GM073678-01 to C. S.-D. Some C. elegans mutant strains were obtained from the Caenorhabditis Genetics Center, which is supported by the NIH National Center for Research Resources (NCRR).
5. Declaration of Interest
None.






