Introduction
Rice (Oryza sativa L.), the staple food crop of the world, is cultivated by the farmers for thousands of years (Fuller, Reference Fuller2011). The variety of rice germplasm on the Asian subcontinent is incredibly diverse and includes landraces, wild Oryza species, natural hybrids between the cultivars and wild relatives and genetic resources derived through various breeding initiatives (Rai, Reference Rai1999). In addition, the southern Himalayan range, Eastern and North Eastern India, Nepal, Myanmar and Thailand are considered to be the areas where domesticated rice originated from its wild predecessors (Chang, Reference Chang1976; Khush, Reference Khush1997; Londo et al., Reference Londo, Chiang, Hung, Chiang and Schaal2006). Additionally, farmers in North Eastern India grow a variety of indigenous rice cultivars under a variety of topographical and agro-climatic conditions. Although the importance of rice diversity in North East India is well recognized (Hore, Reference Hore2005; Durai et al., Reference Durai, Tomar, Devi, Arunachalam and Mehta2015), the evolutionary relationship of indigenous rice landraces in North East India with O. rufipogon and O. nivara is poorly understood. This evolutionary understanding not only assist in knowing the genetic relationship between the wild ancestors and landraces but also helps in proper utilization of distinct primary and secondary gene pools of rice in trait improvement programmes (Brush, Reference Brush1995; Hoisington et al., Reference Hoisington, Khairallah, Reeves, Ribaut, Skovmand, Taba and Warburton1999; Mandel et al., Reference Mandel, Dechaine, Marek and Burke2011). Indigenous rice genotypes are significant sources for evolutionary research specifically in understanding the origin and selection measures involved in fixation of favourable alleles of genes during domestication (Liu et al., Reference Liu, Li and Xing2018). Previous genetic diversity studies using rice genotypes from North East India evaluated the degree of diversity in rice genotypes grown in various seasons, regions and traits such as drought and blast (Choudhury et al., Reference Choudhury, Khan and Dayanandan2013; Das et al., Reference Das, Sengupta, Parida, Roy, Ghosh, Prasad and Ghose2013; Anupam et al., Reference Anupam, Imam, Quatadah, Siddaiah, Das, Variar and Mandal2017; Umakanth et al., Reference Umakanth, Vishalakshi, Sathish Kumar, Rama Devi, Bhadana, Senguttuvel, Sudhir K, Sharma, Sharma, Prasad and Madhav2017; Singh and Singh, Reference Singh and Singh2019). Additionally, nucleotide diversity of domestication genes (OsC1, Wx) in rice genotypes cultivated in North East India has also been assessed (Choudhury et al., Reference Choudhury, Khan and Dayanandan2014).
Since they are inherited uniparentally, mitochondrial and chloroplast genomes are often smaller and structurally distinct to nuclear genomes, with some notable exceptions in plants (Birky, Reference Birky2008). The post-glacial migration routes of variety of plant species has been studied using cytoplasmic markers (cpDNA and mtDNA markers) (Taberlet et al., Reference Taberlet, Fumagalli, Wust-Saucy and Cosson1998; Hewitt, Reference Hewitt2000; Petit et al., Reference Petit, Aguinagalde, de Beaulieu, Bittkau, Brewer, Cheddadi, Ennos, Fineshi, Grivet, Lascoux, Mohanty, MüllerStark, Demesure, Palme, Martin, Rendell and Vendramin2003). The chloroplast DNA has been used preferentially for such studies in plants, because it exhibits a lower rate of nucleotide substitution compared to mitochondrial genome and is less susceptible to extensive intramolecular recombination (Shi et al., Reference Shi, Song, Chen, Cai, Gong, Liu, Shi and Wang2023a, Reference Shi, Song, Liu, Shi and Wang2023b). Further, since there is no recombination in chloroplast genomes, the conserved ancestral genetic structure will be useful not only in understanding the evolutionary relationship using simple phylogenetic studies but also in the proper evolutionary interpretation of the findings (Provan et al., Reference Provan, Powell and Hollingsworth2001). Consequently, the full wild rice chloroplast genome and various genotypes have used to study genetic diversity, population dynamics and evolution.
The chloroplast genome size of indica and japonica rice is around 134 Kb (Tang et al., Reference Tang, Xia, Cao, Zhang, Zeng, Hu, Tong, Wang, Wang, Yu, Yang and Zhu2004). Additionally, Ishii et al. (Reference Ishii, Xu and McCouch2001) used variations in simple sequence repeats found in the rice chloroplast genome to study the diversity of AA genomes. Also, Orf100 and Orf29, two chloroplast markers, have been utilized to distinguish between the genotypes of indica and japonica (Okoshi et al., Reference Okoshi, Nishikawa, Akagi and Fujimura2018). Further, evolution of weedy rice (Oryza sativa f. spontanea), a common weed of rice naturally present in rice fields, has also been studied using chloroplast markers (Yao et al., Reference Yao, Wang, Song, Wang, Liu, Bao and Lu2021). Our previous study has identified that wild relatives and most of the North East landraces were grouped along with Nipponbare (Japonica type) clusters using mitochondrial markers (Parida et al., Reference Parida, Gouda, Chidambaranathan, Umakanta, Katara, Sai, Samantaray, Patra and Mohapatra2023). Therefore, the present study hypothesized that similar relationship might also exist between wild relatives and North East landraces using chloroplast markers. Besides, influence of the chloroplast diversity affecting the evolutionary dynamics of North East rice landraces would also be understood from this study. Specifically, previous studies have not utilized same set of genotypes for chloroplast and mitochondrial diversity analysis (Tong et al., Reference Tong, Kim and Park2016; Cheng et al., Reference Cheng, Nam, Chu, Rungnapa, Min, Cao, Yoo, Kang, Kim and Park2019). Hence, six chloroplast markers showing alignment specific to chloroplast rather than nuclear genome were used to analyse the genetic diversity, population structure and clustering of North East indigenous landraces and wild rice accessions collected from Assam, Manipur and Arunachal Pradesh. This analysis found that North East landraces and wild relatives were grouped with IR64 (indica type) using chloroplast markers rather than Nipponbare cluster.
Materials and methods
Plant materials
A total of 68 landraces including 33 accessions of Assam, 30 from Manipur and five from Arunachal Pradesh were used for evolution study. All the landraces were multiplied in experimental plot of NRRI, Cuttack and observations for yield and domestication-related traits were taken; such as plant height (PH), tiller number (TN), panicle per plant (PPP), panicle length (PL), grains per panicle (GP), length: breadth ratio of grain (LB), panicle type (PT), growth habit (GH), seed shattering (SS), husk colour (HC) and awness (AN). Results of PCA biplot analysis taking PC1 and PC2 showed that all the 68 genotypes were well distributed in that biplot (Supplementary Fig. S1). Also 27 wild and weedy rice accessions including ten O. rufipogon, nine O. nivara and eight weedy rice accessions were used for assessing genetic relatedness. Further, indica rice (IR64) and japonica rice (Nipponbare) were used as reference varieties. However, cultivated varieties are not used in the analysis as it is previously reported to cluster in to indica and japonica types (Singh and Singh, Reference Singh and Singh2019). All rice accessions were grown in NRRI's experimental field during kharif season in 2 years (2015 and 2016 for the period between June and December). Leaf samples were collected after 15 days of transplanting and frozen in liquid nitrogen, stored in −80 °C deep freezer for future use. The detailed information of these genotypes is given in Supplementary Table S1.
Genomic DNA extraction and polymorphism analysis
Plant genomic DNA was collected using a modified CTAB DNA extraction method (Doyle and Doyle, Reference Doyle and Doyle1990) from young leaves that had been frozen in an −80 °C refrigerator. Ground leaf tissue was thoroughly powdered with DNA extraction buffer and incubated for 1 h at 65 °C. Additionally, equal volumes of phenol, chloroform and isoamyl alcohol (25:24:1) were added. The mixture was then centrifuged at 12,000 rpm for 10 min. To completely eliminate the cell debris, the aqueous phase was pipetted out and this process was repeated. The DNA in the aqueous phase was then precipitated using an equivalent volume of isopropanol, and the precipitated pellet was then dried, dissolved in 50 L of TE buffer and stored at −20 °C refrigerator temperature. The quality and quantity of the extracted DNA was analysed using agarose gel electrophoresis along with known amounts of bacteriophage ¥ DNA and NanoDrop 8000 spectrophotometer (Thermo Scientific, Waltham, Massachusetts, USA). Six out of 25 chloroplast-specific markers (4 cp SSR, 2 ORFs) showing 100% alignment only with chloroplast genome of Nipponbare (blast.ncbi.nlm.nih.gov) were selected for this study (Table S2). A total of 50 ng/μl of template DNA was amplified using PCR mixture of dNTPmix (2.5 mM of each), 0.25 μM each of forward and reverse primers, Taq DNA polymerase(0.5 U) and 1X PCR reaction buffer in a total volume of 10 μl in Thermal Cycler (Eppendorf AG, Hamburg, Germany). The initial denaturation was kept at 94 °C temperature, followed by 35 cycles of denaturation at 94 °C for 30s, annealing at 55 °C for 45s, extension at 72 °C for 1 min and a final extension of 10 min at 72 °C. Amplified products were separated on 3.5% MetaPhor™ Agarose gel in horizontal electrophoresis system. Scoring of the amplified bands was done based on the presence and absence of bands or missing data and each band was regarded as an allele and converted to base pair data for further analysis.
Genetic diversity study through statistical analysis and population structure analysis
All the estimates of genetic diversity such as average number of alleles per locus, major allele frequency (MAF), gene diversity, heterozygosity (Ho), polymorphism information content (PIC) and genetic distance were calculated using Power Marker V3.25 (Cavalli-Sforza and Edwards, Reference Cavalli-Sforza and Edwards1967; Liu and Muse, Reference Liu and Muse2005). A phylogenetic tree was constructed based on UPGMA method in PowerMarker using default parameters. Dendroscope V3 was used to visualize the tree (Huson and Scornavacca, Reference Huson and Scornavacca2012). Principal coordinate analysis (PCoA) and AMOVA were performed using GenAlExV6.5 (Peakall and Smouse, Reference Peakall and Smouse2006). Population structure of the rice accessions was analysed using the Bayesian model-based approach employed in STRUCTURE 2.3.4 (Pritchard et al., Reference Pritchard, Stephens and Donnelly2000; Falush et al., Reference Falush, Stephens and Pritchard2003) software. Initially, ten clusters (K) were evaluated using ten replicate runs per K value with a burn-in period length of 10,000 and a run length of 100,000 and a model allowing for admixture and correlated allele frequency. ‘Structure harvester’ programme (http://taylor0.biology.ucla.edu) was used and optimum K value was determined using LnP(D) and Evanno's ΔK (Evanno et al., Reference Evanno, Regnaut and Goudet2005). Population-specific pair-wise values of F-statistics and expected heterozygosity were calculated from the STRUCTURE results.
Results
Genetic diversity analysis of North East accessions using chloroplast markers
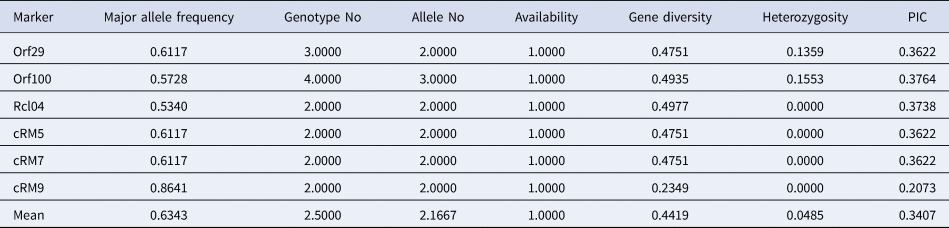
Among all the chloroplast primers used in this study, five primers amplified two numbers of alleles per locus and only a single primer, ORF100 amplified three alleles per locus. The average value of 2.166 alleles per marker was detected in selected rice genotypes. Further, PIC value obtained using chloroplast-based primers ranged from 0.207 (cr09) to 0.376 (ORF100) with an average value of 0.340. Besides, MAF ranged between 0.534 (Rcl04) and 0.864 (cr09), and gene diversity (G) ranged between 0.234 (cr09) and 0.497 (Rcl04). Further, heterozygosity (Ho) ranged from 0.0 (Rclo4, cr05, cr07, cr09) to 0.155 (ORF100) for chloroplast primers in the North East accessions (Table 1).
Table 1. Genetic diversity and PIC of chloroplast markers using NE accessions
Hierarchical cluster analysis using chloroplast markers
The reference genotypes IR64 and Nipponbare were genetically grouped into two well-separated clusters in UPGMA tree (cluster I: Nipp cluster and cluster II: IR64 cluster). Rice accessions from North Eastern states of Assam and Manipur were equally distributed in both indica (IR64) and japonica (Nipponbare) major clusters. The major cluster I comprised of 19 Assam rice and 15 Manipur rice accessions along with japonica cultivar Nipponbare. The second major cluster II comprised of 14 Assam rice and 15 Manipur rice accessions and indica cultivar IR64. Besides, all the five Arunachal Pradesh accessions were grouped in Nipponbare major cluster I (Fig. 1). The list of genotypes in different clusters is given in Table S3. The AMOVA analysis revealed a higher-level variation among individuals (84%) than among the populations (5%) and within individuals (11%) (Fig S2). The allelic pattern of the genotypes is given in Fig. S3.
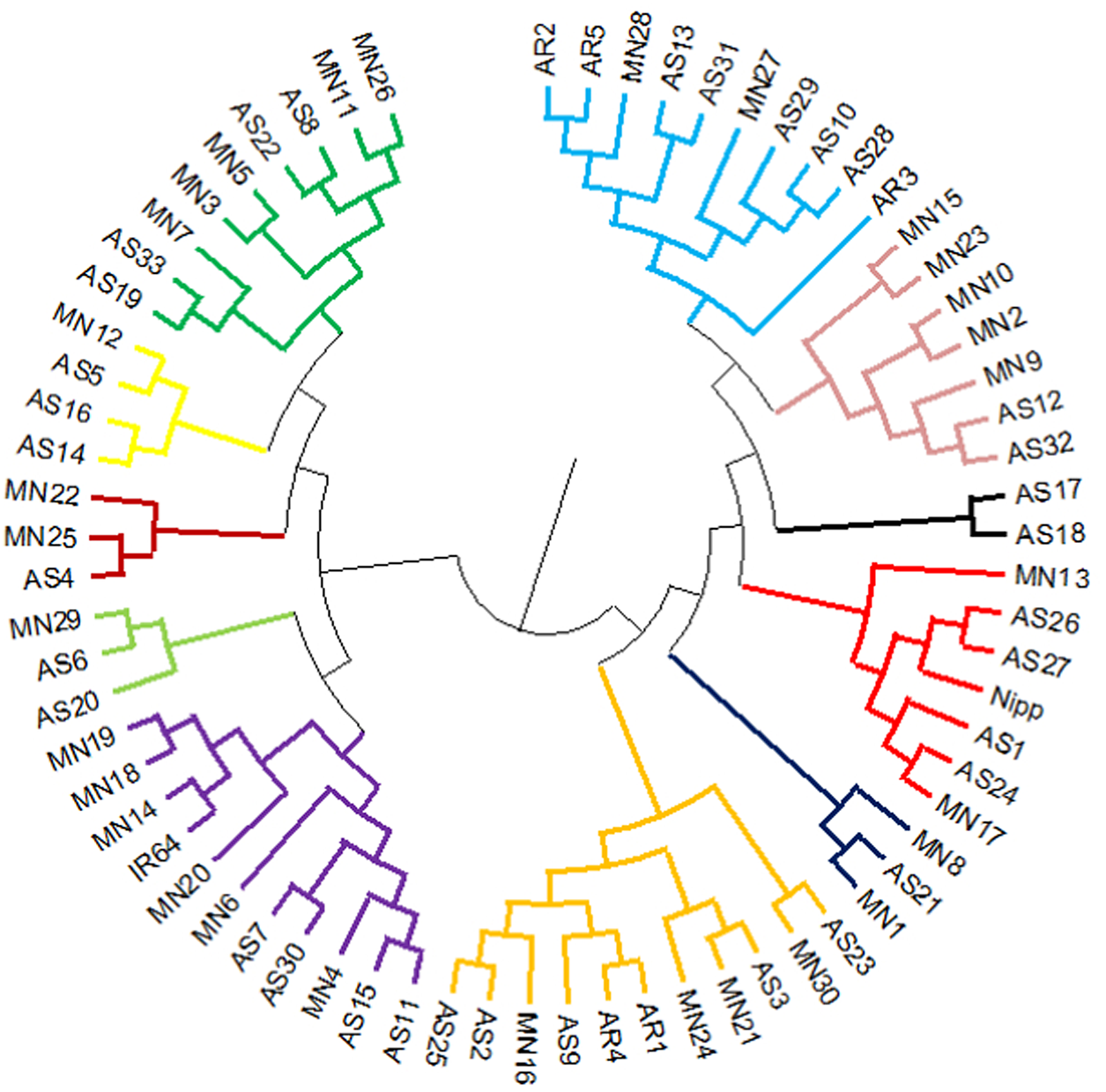
Figure 1. Cluster diagram of 68 North East landraces and two check varieties (IR64 and Nipponbare) using six chloroplast markers. AS, Assam; AR, Arunachal Pradesh; MN, Manipur.
Population structure analysis using chloroplast markers
Two sub-populations (K2) were present in the studied accessions using the chloroplast markers (Fig. 2). Further, sub-population I comprised of 13 Assam rice, 12 Manipur rice and five Arunachal Pradesh rice accessions. Similarly, 14 Assam rice and 14 Manipur rice accessions along with IR64 were grouped in sub-population II. Most of the wild rice collections were grouped with IR64 subgroup. In addition, six Assam rice and four Manipur rice were grouped as admixtures along with Nipponbare. However, O. rufipogon and O. nivara were grouped in separate sub-populations in the barplots of structure analysis using K3–K6. A similar pattern was also observed for the landraces of Assam, Manipur and Arunachal Pradesh.

Figure 2. Structure analysis of North-East rice accessions with two check varieties using chloroplast markers.AS, Assam; AR, Arunachal Pradesh; MN, Manipur.
PCoA and pair-wise genetic diversity analysis
The PC1 component showed the highest proportion of variance (35.74%), followed by the second and third axes (19.58 and 18.85%, respectively) with cumulative variation of 55.32% (Fig. S4). The PC1 components contained 15 Assam rice, 17 Manipur rice, five Arunachal Pradesh rice and Nipponbare, whereas PC2 components contained 18 Assam rice, 13 Manipur rice and five Arunachal Pradesh rice and Nipponbare. Further, three Manipur rice accessions (MN14 (AC9211), MN18 (AC9242) and MN19 (AC9246)) were grouped with IR64 coordinates and two Assam accessions (AS26 (AC35726) and AS27 (AC35727)) were grouped with Nipponbare coordinates. Further, Nipponbare and North East accessions, as well as IR64 and North East accessions, were found to have genetic distances between them of 0.47 and 0.55, respectively (Fig. S5).
Genetic diversity analysis of wild and weedy rice using chloroplast markers
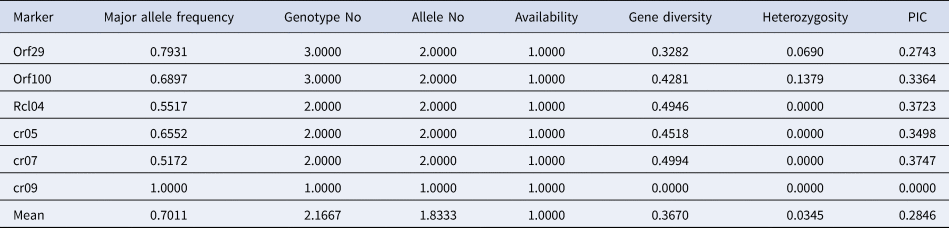
Five of the six chloroplast markers that were used to analyse the genotypes of wild and weedy rice amplified two alleles per gene, while the cr09 marker amplified just one allele per allele. Similar to North East accessions, the investigated wild and weedy rice had an average value of 2.166 alleles per marker. The PIC values for the chloroplast markers were also generated, and the range was 0 (cr09) to 0.374 (cr07), with an average value of 0.284. Additionally, gene diversity (G) ranged between 0 (cr09) and 0.499 (cr07), while MAF ranged from the lowest value of 0.517 (cr07) to the highest value of 1 (cr09). In wild and weedy rice, the chloroplast marker alleles displayed heterozygosity (Ho) ranging from 0.0 (Rclo4, cr05, cr07, cr09) to 0.137 (ORF100) (Table 2).
Table 2. Genetic diversity and PIC of chloroplast markers in wild and weedy rice
Cluster analysis of wild and weedy rice along with IR64 and Nipponbare using chloroplast markers
Two major clusters were identified in the wild and weedy rice genotypes using six chloroplast markers (Fig. 3). The major cluster I comprised of a wild rice (Niv5) and four weedy rice (WD3, WD5, WD6, WD7) genotypes. The second major cluster comprised of the remaining 18 wild rice, four weedy rice and IR64 and Nipponbare reference cultivars (Table S4 and Fig S6). The cluster analysis with North East landraces showed five landraces of Arunachal Pradesh, five wild and weedy rice genotypes, 19 Assam rice landraces, the japonica check (Nipponbare) and 14 Manipur landraces made up the major cluster I. Intriguingly, the majority of the wild and weedy rice (22 nos.) as well as the indica check cultivar IR64 made up the second major cluster, which also included the remaining 14 numbers of Assam rice, 14 Manipur accessions and other rice varieties (Fig. 4, Table S5).
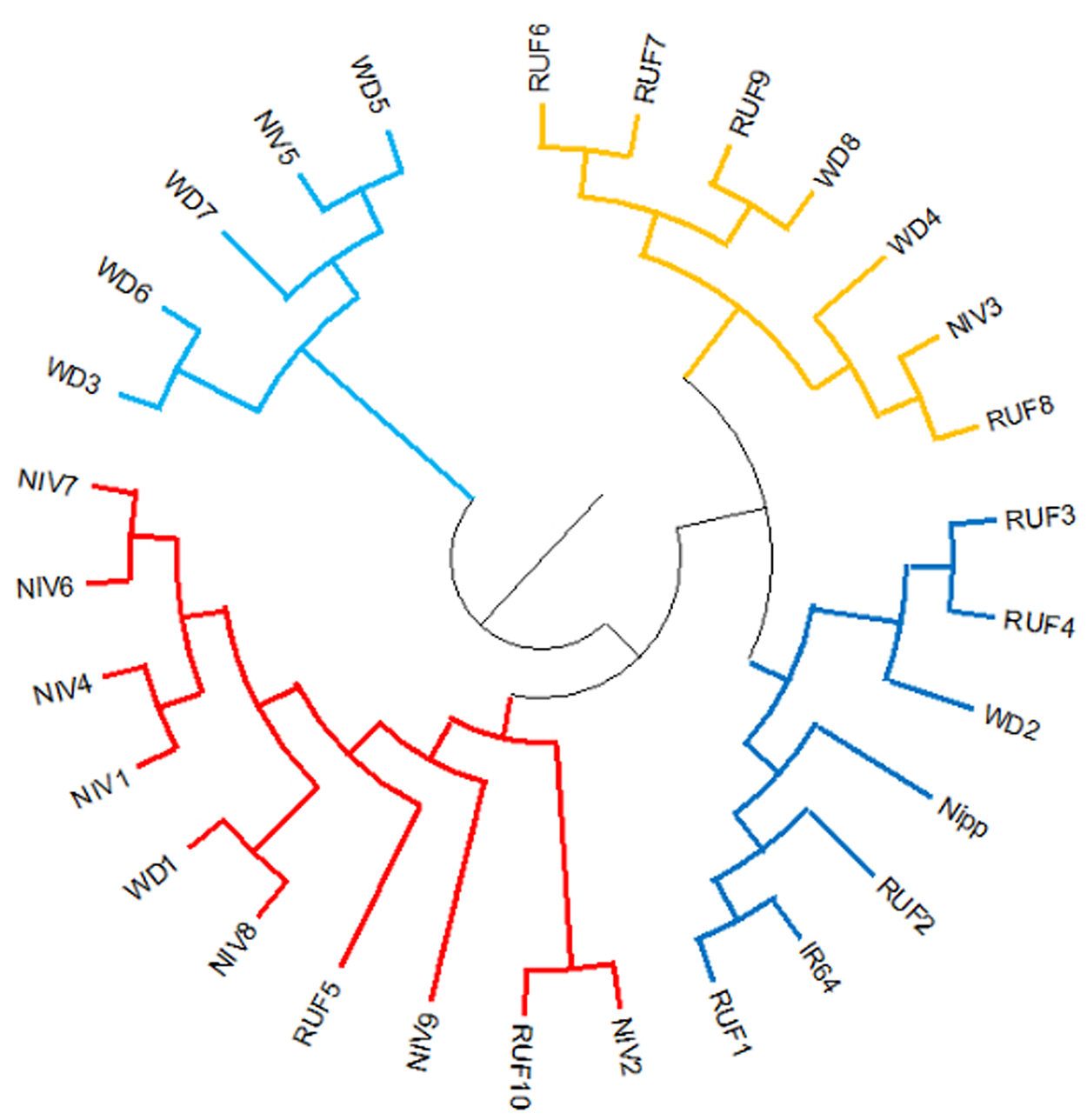
Figure 3. Cluster diagram of wild and weedy rice and two check varieties (IR64 and Nipponbare) using six chloroplast markers. RUF, O. rufipogon; NIV, O.nivara; WD, weedy rice.
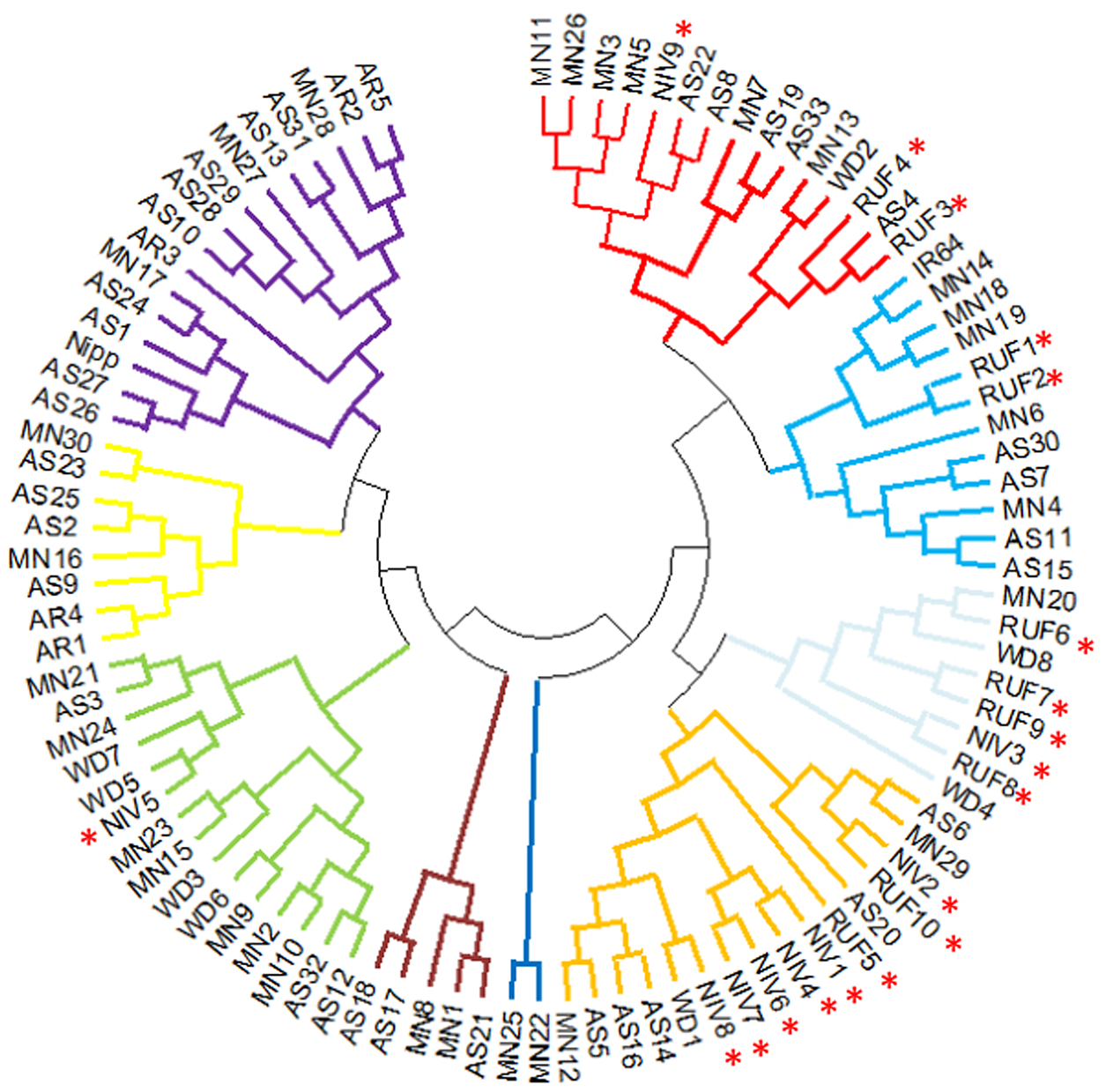
Figure 4. Cluster diagram of NE accessions, wild and weedy rice and two check varieties (IR64 and Nipponbare) using six chloroplast markers. Wild rice accessions are highlighted using star symbol. RUF, O. rufipogon; NIV, O.nivara; WD, weedy rice; AS, Assam; AR, Arunachal Pradesh; MN, Manipur.
Discussion
The evolutionary relationships and genetic diversity of North East rice accessions are poorly understood. A proper knowledge on genetic relationship assists us in better utilization of distinct rice gene pools in crop improvement programmes. To address this, we used chloroplast markers to investigate the diversity of chloroplasts and the evolutionary relationship between North East accessions and the wild relatives of rice. Two of the six markers under investigation (Orf100 and Orf29) showed the allelic difference between IR64 and Nipponbare. In agreement with earlier studies (Chen et al., Reference Chen, Nakamura, Sato and Nakai1993; Li et al., Reference Li, Kang, Zhang, Huang and Chen2012a, Reference Li, Zhang, Huang, Kang, Liang and Chen2012b) the deletion in both the Orf100 and Orf29 markers was distinct and present in the majority of indica genotypes and these allelic differences were used to group the landraces into distinct indica and japonica subtypes. Two chloroplast markers (Orf29, Orf100) also showed heterozygosity in the range of 0.1359–0.1553 in our analysis. In support of our observation, chloroplast variation map generated using re-sequencing methodology in Korean rice landraces also identified ~23% of variation was due to heterozygosity in chloroplast genomes (Tong et al., Reference Tong, He, Wang, Yoon, Ra, Li, Yu, Oo, Min, Choi, Heo, Yun, Kim, Kim, Lee and Park2015). According to Waters et al. (Reference Waters, Nock, Ishikawa, Rice and Henry2012), chloroplast genomes are typically circular haploids, and the presence of heterozygous alleles denotes the existence of diverse chloroplast subtypes and was attributed to recent bottleneck events or strong selection (Cheng et al., Reference Cheng, Nam, Chu, Rungnapa, Min, Cao, Yoo, Kang, Kim and Park2019). Therefore, further research is needed to determine whether heterozygous alleles found in ten North East accessions also point to similar selection pressure or bottleneck events.
Our results indicate that a high level of genetic diversity in chloroplast genomes pre-existed in the ancestral pool of North East accessions. The mean genetic diversity and PIC of North East accessions were 0.4419 and 0.3407, respectively. Our results are in agreement with those of Ishii et al. (Reference Ishii, Xu and McCouch2001) who reported that the mean PIC value of 29 cultivars and 30 accessions of AA genomes was also 0.38. Also, Kim et al. (Reference Kim, Jeong, Ahn, Doyle, Singh, Greenberg, Won and McCouch2014) showed that the mean gene diversity of a mini rice diversity panel made up of tropical and temperate japonica, indica, aus and aromatic ecotypes was only 0.202. Therefore, ecological adaptation of landraces unique to the agroclimatic conditions in North East India needs to be further studied.
In agreement with previous findings, two distinct sub-populations specific to indica and japonica groups were identified in our analysis. Previous studies using complete chloroplast genome sequences for substructure determination also clearly showed that the indica and japonica genotypes were divided into separate subpopulations (Tong et al., Reference Tong, Kim and Park2016; Cheng et al., Reference Cheng, Nam, Chu, Rungnapa, Min, Cao, Yoo, Kang, Kim and Park2019). However, in contrast, distinct indica and japonica subpopulations were not found in cultivars from the Philippines, Pakistan and India using both chloroplast and mitochondrial markers (Shah et al., Reference Shah, Aslam, Shabir, Khan, Abbassi, Shinwari and Arif2015). This distinct grouping of geographically limited sampling regions of North East India in this analysis indicates the genetic diversity for both indica and japonica gene pools in chloroplast genome. Similarly, North East landraces were split into two main clusters. A phylogenetic analysis performed by Garris et al., (Reference Garris, Tai, Coburn, Kresovich and McCouch2005) and Moner et al. (Reference Moner, Furtado and Henry2020) using the chloroplast genome also showed two distinct clades among the approximately 3091 different rice ecotypes, supporting our observation. Further, two clusters found in North East rice accessions may indicate two distinct and independent evolutionary trajectories present in the studied accessions for chloroplast markers (Cheng et al., Reference Cheng, Nam, Chu, Rungnapa, Min, Cao, Yoo, Kang, Kim and Park2019).
Wild rice and North East accessions were divided into two main clusters. In contrast to the Nipponbare cluster, the majority of the wild rice genotypes were grouped in IR64 cluster along with majority of landraces using chloroplast markers. This finding is in sharp contrast to our previous findings that most of the wild relatives and landraces were grouped in Nipponbare cluster using mitochondrial markers (Parida et al., Reference Parida, Gouda, Chidambaranathan, Umakanta, Katara, Sai, Samantaray, Patra and Mohapatra2023). This indicates the maternal and paternal ancestors are distinct for the two main groups of rice landraces in North East India. In earlier studies, Li et al. (Reference Li, Kang, Zhang, Huang and Chen2012a, Reference Li, Zhang, Huang, Kang, Liang and Chen2012b) used chloroplast markers such as Orf100 and Orf29 and identified both indica and japonica as having originated from various O. rufipogon strains. Furthermore, Tong et al. (Reference Tong, Kim and Park2016) demonstrated that japonica and indica originated from O. rufipogon and O. nivara, respectively. Using genome-wide chloroplast variants, Wambugu et al. (Reference Wambugu, Brozynska, Furtado, Waters and Henry2015), Cheng et al. (Reference Cheng, Nam, Chu, Rungnapa, Min, Cao, Yoo, Kang, Kim and Park2019) and Gao et al. (Reference Gao, Liu, Zhang, Li, Gao, Liu, Li, Shi, Zhao, Zhao, Jiao, Mao, Gao and Eichler2019) also reported findings of a similar nature. Contrary to these reports, only a small number of accessions (MN12, AS16, AS14 and AS1; MN12 (AC9139), AS16 (AC35715), AS14 (AC35713) and AS1 (ARC10322)) were specifically clustered with O. nivara in our analysis. This also suggests that the North East accessions’ chloroplast genomes may have originated from various O. rufipogon and O. nivara ancestor pools. The unique clustering pattern of North East landraces with O. rufipogon and O. nivara suggests a likely distinct evolutionary origin and contribution of both the wild relatives in the origin of domesticated rice in North East India. Besides, not only the wild relatives, but also the maternal and paternal wild parents would have been distinct between the two groups of rice landraces in North East India.
Conclusion
North East India represents rich genetic diversity region for rice in India. Six markers in the chloroplast genomes were analysed, and two major clusters representing the reference genotypes IR64 and Nipponbare were found in the North East landraces of Assam, Manipur and Arunachal Pradesh. Furthermore, 43% of the studied North East rice accessions were found in IR64 cluster, which also contained the majority of the wild rice genotypes. The likelihood of different wild ancestors, maternal and paternal ancestors, and distinct domestication events leading to two phylogenetic clusters of rice in North East India is indicated by this analysis. This study provides important new information about the diversity of North East landraces for chloroplast markers, and future work using complete chloroplast genome sequences may help to understand the evolutionary pattern connected to rice domestication events in North East India.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S1479262123000990.
Acknowledgements
We gratefully acknowledge the financial support provided by the Indian Council of Agricultural Research (ICAR), New Delhi for successfully completing this work. Also, we sincerely acknowledge the Director, National Rice Research Institute (ICAR-NRRI), Cuttack for providing necessary lab facilities for completion of this work.
Author contributions
Conceptualization of the work: Trilochan Mohapatra; collection of materials for research: Bhaskar Chandra Patra, Ngangkham Umakanta; genotyping and PCR: Madhuchhanda Parida; data analysis: Madhuchhanda Parida, Gayatri Gouda, Parameswaran Chidambaranathan; finalization of analysis: Trilochan Mohapatra, Paramswaran Chidambaranathan; manuscript draft writing: Madhuchhanda Parida and Cayalvizhi Balasubramania Sai; manuscript editing and finalization: Madhuchhanda Parida, Parameswaran Chidambaranathan, Sanghamitra Samantaray; overall coordination: Trilochan Mohapatra.








