INTRODUCTION
Campylobacteriosis is a leading zoonotic foodborne disease in Europe and around the world. Its latest yearly incidence in Europe and Slovenia is 50.28 and 48.7/100 000, respectively [1, 2]. These numbers are still believed to be strongly underreported, since most campylobacteriosis cases remain unrecognized by surveillance systems due to the mild self-limiting symptoms or undetermined cases of gastroenteritis. In Slovenia, as well as in other countries, there is a trend of increasing campylobacteriosis incidence [2, 3].
Of the thermotolerant campylobacters, Campylobacter jejuni is the most infectious, causing around 90% of reported human campylobacteriosis cases in Slovenia [2]. Besides the high prevalence of infections, antibiotic resistance of C. jejuni against quinolones (ciprofloxacin and enrofloxacin) and to a lesser extent, macrolides (erythromycin and azithromycin) is problematic [Reference Kurinčič, Klančnik and Smole4]. In Slovenia in 2010, 78% (n = 60) of C. jejuni strains isolated from meat were resistant against ciprofloxacin, while all of them were susceptible to erythromycin. Of the human C. jejuni clinical isolates from the same year, 67.2% and 1.0% (n = 882) were resistant against ciprofloxacin and erythromycin, respectively [3]. This degree of resistance and the emergence of multidrug-resistant strains can seriously hinder clinical treatment of campylobacteriosis [Reference Smole5].
Campylobacter colonizes the gut of many production animals, especially broilers, in which it rarely causes disease symptoms. The meat of these animals is therefore often contaminated during slaughtering and serves as a vehicle of transmission of this pathogen via the food production chain [Reference Silva6]. Campylobacter is widely known for its rapid adaptive ability and genomic instability [Reference Jayaraman7–Reference Wassenaar, Geilhausen and Newell9]. When different reservoirs harbour strains with specific genotypes, it is possible to determine the source of the infection by pulsed-field gel electrophoresis, multilocus sequence typing (MLST) and/or fla gene typing [Reference Sahin10–Reference Sheppard12]. Nevertheless, MLST has become the method of choice for Campylobacter genotyping. Moreover, in combination with sequencing of the short variable region within the flagella-encoding flaB gene MLST allows further strain differentiation within the same sequence type (ST) [Reference Wirz13]. This approach has allowed the correlation of specific genotypes with specific antimicrobial resistance profiles [Reference Wirz13, Reference Gu14]. It was shown that resistance to quinolones and macrolides are mostly associated with point mutations in gyrA and 23S rRNA, respectively, acting together with efflux mechanisms [Reference Kurinčič, Klančnik and Smole15, Reference Kurinčič16]. The RND-type efflux pumps of some Gram-negative bacteria are able to extrude different types of antibiotics and can also be induced by their substrates [Reference Piddock17]. This indicates that environmental selective pressures play an important role in acquiring non-specific (via active efflux), or specific (via point mutation), resistance against antimicrobial drugs. Specific STs have already been associated with quinolone resistance in strains with point mutations, but it is not yet known whether this is because these genotypes are more prone to mutations conferring resistance or because they are clonal [Reference Wirz13].
We investigated the potential correlation between MLST/flaA genotypes, source of the isolates and resistance against seven antibiotics of 52 Slovenian C. jejuni isolates, in order to elucidate the spread of antibiotic resistance. This is the first report on MLST/flaA characterization of Slovenian C. jejuni isolates from humans, animals, water, and food.
MATERIALS AND METHODS
Campylobacter strains and growth conditions
Fifty-two C. jejuni strains isolated from different sources (human, raw and cured chicken meat from retail, animal, water) and regions in Slovenia were used in our study. Control reference strains included C. jejuni ATCC 33 560 and C. coli strains 158 and 01378 as negative controls. The majority of meat and animal isolates, and all water isolates from Slovenian rivers were obtained through national monitoring in 2008/2009. Five chicken meat isolates were obtained from a laboratory collection and human isolates were from reported campylobacteriosis cases in 2009. C. jejuni was confirmed by hipO gene species-specific polymerase chain reaction (PCR) [Reference Zorman and Smole18]. Isolates were grown on Columbia blood agar (Oxoid, UK) under microaerobic conditions (5% O2, 10% CO2, 85% N2) at 42°C for 24 h and maintained at −80°C in brain heart infusion broth (Oxoid) with 5% horse blood (Oxoid) and 20% glycerol.
MLST and flaA short variable region sequence typing
DNA was extracted from 24-h cultures with PrepMan® (Applied Biosystems, USA) according to the manufacturer's instructions and MLST was performed as described by Dingle et al. [Reference Dingle19]. Seven housekeeping genes, aspA, glnA, gltA, glyA, pgm, tkt, uncA were amplified using Taq polymerase (Promega, USA) with primers detailed in Korczak et al. [Reference Korczak20]. The short variable region of the flaA gene (flaA-SVR) was amplified by primers flaA_Cjc-L and flaAB_Cjc-R [Reference Korczak20] according to the protocol of Josefsen et al. [Reference Josefsen21]. The purified PCR products were sequenced with the BigDye terminator v. 3.1 ready reaction cycle sequencing kit (Applied Biosystems) on an ABI3130XL genetic analyser (Applied Biosystems).
The sequences of the seven MLST loci and flaA were compared with sequences in the MLST database (www.pubmlst.org/campylobacter) to determine the allele number. The STs, clonal complexes (CCs) and flaA type of each strain were assigned from the profiles of the seven alleles in the MLST database with an integrated automated link in the computer program BioNumerics v. 6.6 (Applied Maths NV, Belgium). Relationships between strains were determined by the MLST-based network creation method, minimum spanning tree (MST) and by the unweighted pair-group method with arithmetic mean cluster analysis based on flaA-SVR in BioNumerics. New alleles and STs were submitted to the PubMLST database.
Antibiotic susceptibility
The susceptibility of isolates to seven antibiotics (chloramphenicol, ciprofloxacin, erythromycin, gentamicin, nalidixic acid, streptomycin, tetracycline) was determined by a broth microdilution method using a commercial diagnostic test for Campylobacter minimum inhibitory concentrations (MICs) (Sensititre Campylobacter plate – EUCAMP; Trek Diagnostic Systems, USA) according to the manufacturer's instructions. Resistance against antibiotics was expressed as MICs based on cut-off values recommended by European Food Safety Authority (EFSA) [22]. Multidrug resistance was defined as resistance to at least three non-related antibiotics.
Mutation assay PCR and gyrA sequencing
Mismatch amplification mutation assay (MAMA) PCR for detection of the Thr86-Ile mutation, which confers resistance against ciprofloxacin was performed on all phenotypically ciprofloxacin-resistant isolates. Additionally, the quinolone resistance-determining region (QRDR) of gyrA was sequenced (Macrogen, South Korea) to study the genetic relatedness of ciprofloxacin-resistant isolates from different CCs. Both procedures were as described previously [Reference Zirnstein23]. Thirty-two sequences of gyrA QRDR were deposited in GeneBank (accession numbers KF831198–KF831229).
Statistical analysis
The discriminatory index of the typing methods was calculated by Simpson's index of diversity according to Hunter & Gaston [Reference Hunter and Gaston24]. Statistical significance (P < 0.05) of association of specific genotypes with quinolone resistance and source of the strains was tested using IBM SPSS Statistics v. 20 (IBM, USA), with Fisher's exact two-tailed test and Pearson's χ 2 test.
RESULTS AND DISCUSSION
The 52 C. jejuni isolates were recovered from animals (chicken, bovine, turkey skin and faeces), raw and cured chicken meat from retail, human patients and environmental water samples during 2008–2009 in different regions of Slovenia. The isolates were grouped by MLST into 28 STs, four of which were novel additions to the PubMLST database; one of these new STs (ST5207 – C. jejuni 1603) was a novel combination of previously known alleles but three, ST5205, ST5206, ST6309, included novel allele sequences in tkt, aspA and glyA loci, respectively. Each of the isolates with the novel tkt allele type originated from the same region in Slovenia (data not shown) and were isolated from raw and cured chicken meat and human faeces, which is suggestive of chicken being the most probable source of infection.
As shown in Table 1, 48 C. jejuni isolates were assigned to ten pre-defined CCs and 24 STs by MLST. Four isolates could not be assigned to a CC. CC21 and CC45 predominated and accounted for 19 and eight, respectively, of the total 52 isolates typed. ST50 and ST104 were the most frequent STs accounting for 10 and six, respectively, of the total (Fig. 1, Table 1). Twenty-three (79%) STs appeared only once, indicating high genetic diversity. STs belonging to CC21 and CC45 were previously shown to be predominant in Europe and typical for poultry [Reference Wirz13, Reference Korczak20, Reference Gripp25, Reference De26]. In our study, 12/19 isolates assigned to CC21 and 6/8 assigned to CC45 were isolated from poultry meat or faeces.

Fig. 1. Phylogenetic tree constructed on the basis of the short variable repeat of the flaA region showing correlation between antibiotic resistance, origin of the strains and MLST type. Light grey squares represent antibiotic-sensitive phenotypes; dark grey squares represent antibiotic-resistant phenotypes; black squares represent ciprofloxacin-resistant phenotypes of clonal complex ST21. CC, Clonal complex; ST, sequence type; ND, not determined.
Table 1. Prevalence of Campylobacter jejuni isolates from different sources in identified clonal complexes in this study

CC, Clonal complex; STs, sequence types.
The distribution of isolates in the MST analysis (Fig. 2) indicates some correlation between specific genotypes and their main source of isolation. The majority (71%) of human isolates was distributed among CC21 and CC353 and most (67%) of the meat isolates among CC21 and CC45, while animal isolates were mostly (67%) assigned to CC21 and CC354. Water isolates were genetically the most diverse group and 2/5 could not be assigned to any existing CC. Their distinct STs present an interesting group with little in common with isolates from human and animal sources. Water as an abiotic source, represents a different environment to which bacteria need to adapt phenotypically and genetically. Similar genetic differences between C. jejuni strains originating from biotic and abiotic sources have been found in the past and resulted in the identification of specific genetic markers able to distinguish between isolates from biotic and abiotic sources [Reference Champion27].
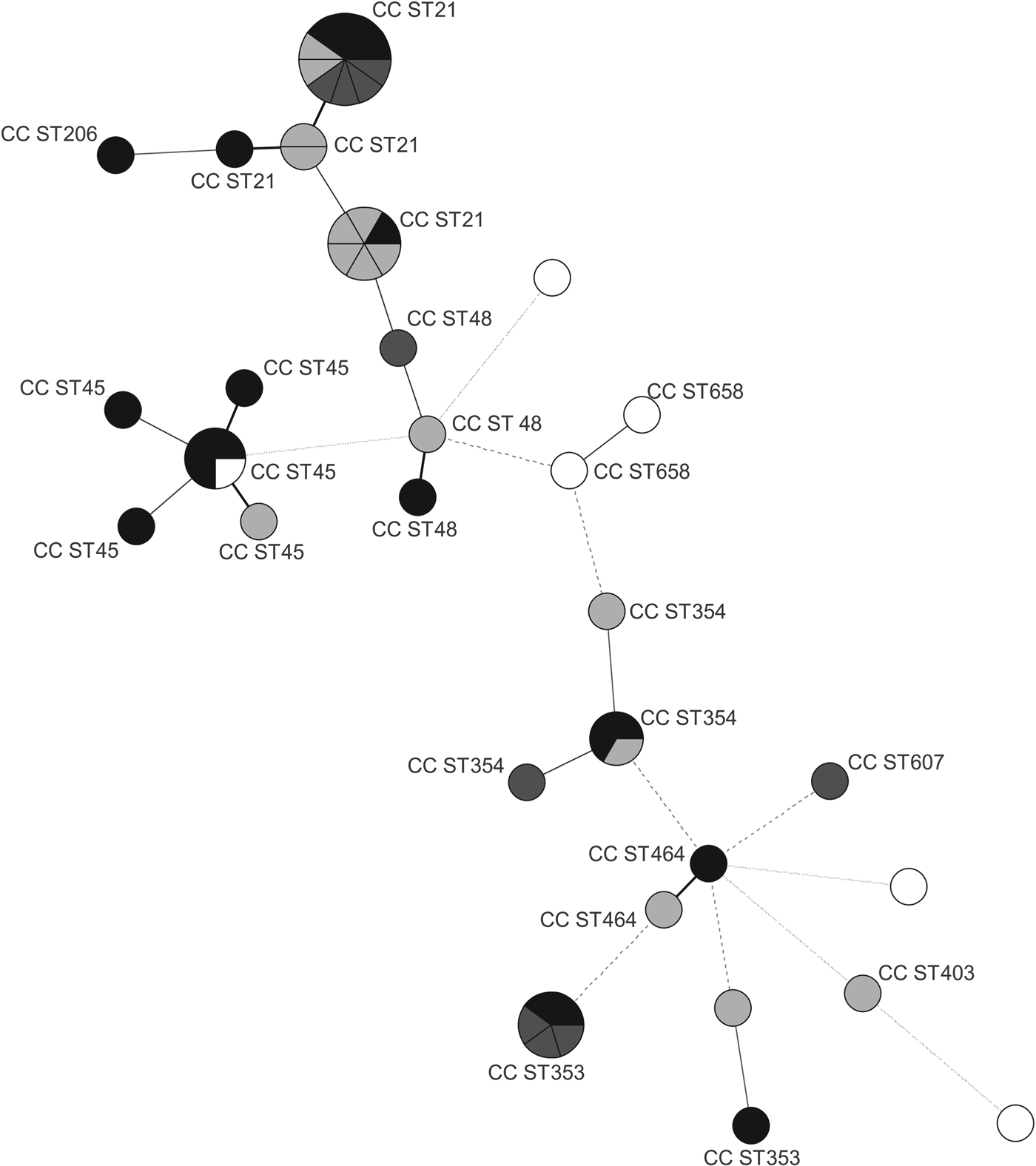
Fig. 2. Minimum spanning tree generated from MLST comparisons of Campylobacter jejuni strains isolated from animals, humans, meat, and water in Slovenia. Each circle represents one sequence type (ST), the clonal complexes (CC STs) are indicated by numbers. Node sizes represent higher strain numbers within one ST. Node colour indicates the origin of the strains: black (meat), dark grey (human), light grey (animal), white (water). The connecting lines between STs depict the number of allelic differences between them: one allele difference (black bold lines), two alleles difference (grey bold lines), three alleles difference (grey dashed lines), more than three alleles difference (grey dotted lines).
flaA-SVR typing revealed 22 different genotypes in the C. jejuni isolates (Fig. 1), and was thus a less discriminatory method than MLST (28 STs). Indeed, flaA-SVR typing additionally discriminated isolates within only one ST (ST50), whereas MLST distinguished between isolates in more flaA types. This was reflected by the higher discriminatory power of MLST (0.942) over flaA-SVR (0.928). The combination of both typing methods increased the discriminatory power to 0.967.
The most common flaA alleles were 36, 278 and 256 with prevalences of nine (17%), eight (15%) and seven (13%) out of a total of 52 alleles. All flaA allele types 256 and 36 were segregated into CC21, while most of the strains with allele type 278 were assigned to CC353, as well as to CC21 and CC48. Cluster analysis of flaA-SVR sequences revealed six major clusters at a similarity level of 95%. Statistical analysis confirmed flaA allele type 36 (8/9) as typical for animal isolates and type 278 (5/8) as typical for meat isolates.
The highest frequency of antibiotic resistance was recorded for the quinolones ciprofloxacin and nalidixic acid (61% and 58%), followed by tetracycline (25%), streptomycin and erythromycin (4%) (Table 2). All C. jejuni isolates were susceptible to gentamicin and chloramphenicol. These results are in accordance with trends reported by the EFSA [3]. Antibiotic resistance in C. jejuni is becoming a significant issue, not only in Slovenia, but also in other European countries [3] with the most problematic being resistance against quinolones, which are the second group of drugs of choice for campylobacteriosis treatment. In Slovenia quinolones are widely used for the non-specific treatment of bacterial gastroenteritis as well as for enterococcal and E. coli infections in poultry flocks. The prevalence of ciprofloxacin-resistant animal Campylobacter isolates in Europe is reported to range from 37% to 84%, depending on the country [3] which is consistent with the results reported here, where 72% of animal and 44% of meat isolates were ciprofloxacin resistant and this increased to 80% for human isolates. Almost all (94%) ciprofloxacin-resistant strains from our study were cross-resistant to nalidixic acid. Interestingly, tetracycline resistance occurred almost exclusively (92%) in ciprofloxacin-resistant strains. Although erythromycin resistance was rare, it is of concern that both erythromycin-resistant isolates were from raw chicken meat and exhibited multidrug resistance to ciprofloxacin, nalidixic acid, streptomycin and tetracycline, which seriously compromises the choice of treatment for infections caused by the ingestion of these organisms.
Table 2. Distribution of antibiotic resistant strains in clonal complexes

CC, Clonal complex; STR, streptomycin; CIP, ciprofloxacin; TET, tetracycline; ERY, erythromycin; NAL, nalidixic acid; MDR, multidrug-resistant strain (resistant against ⩾3 unrelated antibiotics).
The high incidence of quinolone resistance compared to erythromycin resistance was previously demonstrated to be due to the faster development of ciprofloxacin-resistant mutants during exposure to antibiotics, improved biological fitness of resistant mutants and their ability to persist in the environment after the selective pressure is removed [Reference Price28–Reference McDermott30]. By contrast, macrolide resistance appears to require longer exposure to antibiotics to develop, and resistant strains are less likely to survive in the absence of selective pressure [Reference Luangtonkum31–Reference Luangtongkum33]. We also observed persistence of ciprofloxacin-resistant strains of the same genotypes (e.g. ST50) which were isolated from animals, meat, and human samples.
Analysis of antibiotic resistance and MLST genotypes revealed that all but one of the 19 strains of C. jejuni belonging to the predominant CC21 were quinolone resistant (P < 0.05). Over half of ciprofloxacin-resistant genotypes within CC21 belonged to ST50 and over 30% to ST104 and only 13 (42%) of these resistant strains were identified in all other CCs (P < 0.05). A similar pattern was observed with flaA types, where 47% of ciprofloxacin-resistant CC21 genotypes were assigned to flaA 36 and 32% to flaA 265. All ciprofloxacin-resistant strains in CC21 also showed cross-resistance to nalidixic acid. An association between CC21 and quinolone resistance has already been found in Belgium [Reference Habib34] and Switzerland [Reference Wirz13] accounting for resistance rates in these countries of 66% and 30%, respectively.
In order to determine whether the association of quinolone resistance with CC21 was due to individual mutational events or an increased ability to spread clonally [Reference Hunter and Gaston24], we investigated the most common ciprofloxacin resistance conferring mutation Thr86-Ile and analysed the QRDR by sequence typing. The results show that all phenotypically ciprofloxacin-resistant isolates, with the exception of strain 180/08, have the mutation ACA to ATA in the 86th codon. Six additional silent mutations in the QRDR were identified, and on the basis of these results we constructed the dendrogram shown in Figure 3, which clearly indicates the high degree of genetic relatedness of ciprofloxacin-resistant isolates from CC21, which together with one strain from CC658 and two strains with unassigned CCs form a separate group compared to the ciprofloxacin-resistant strains from all other CCs. As quinolone resistance is quickly developed under antibiotic selective pressure and can persist long after cessation of treatment, we conclude that the high incidence of such strains within CC21 found here is not due to high genetic plasticity of this particular CC [Reference Hunter and Gaston24], but rather to acquired efficiency of clonal spreading. This hypothesis is additionally supported by the fact that not all investigations from other countries [Reference Wang35] found a correlation between quinolone resistance and CC21.

Fig. 3. Relationship between Campylobacter jejuni strains based on the sequences of quinolone resistance-determining region (QRDR) of the gyrA gene in comparison to clonal complex (CC). The position of silent mutations in relation to the reference strain C. jejuni NCTC 11158 are shown on the branches. The accession numbers of the sequences are shown in parentheses after the strain designation. ND, Not determined.
In conclusion, this study is the first report on the genetic variability of C. jejuni isolates from Slovenia based on MLST, which contributes new allele types and STs to the PubMLST database. CC21 was the predominant genetic group with representative isolates from a variety of different sources and this group was highly associated with quinolone resistance, confirming its clonal spreading along the food supply chain. We also identified distinct genotypes of strains from water sources with high antibiotic susceptibility.
ACKNOWLEDGEMENTS
This study was partly co-financed by PROMISE (FP7-265877), Biotracer (FP6-036272) and CRP V4-1110. The authors thank Jonas Larsson and Daniela Petek for their contribution with sequencing and sequence analysis.
DECLARATION OF INTEREST
None.








