1. Introduction
Genetic distortion or segregation deviating from Mendelian ratios is not a rare case in segregated populations. Segregation distortion is the loss of specific genotypes in progeny, which was caused by gametic or zygotic selection due to a number of physiological or genetic factors, such as competition among gametes or pollens for preferential fertilization (Wendel et al., Reference Wendel, Edwards and Stuber1987; Lyttle, Reference Lyttle1991) and sexual-reproductive sterility due to inbreeding depression (Remington & O'Malley, Reference Remington and O'Malley2000). It was correlated with increasing genetic divergence between parental lines (Paterson et al., Reference Paterson, Damon, Hewitt, Zamir, Rabinowitch, Lincoln, Lander and Tanksley1991; Grandillo & Tanksley, Reference Grandillo and Tanksley1996; Bradshaw et al., Reference Bradshaw, Otto, Frewen, McKay and Schemske1998). Segregation distortion, which is considered to be a major genetic factor of divergence (McDaniel et al., Reference McDaniel, Willis and Shaw2007), may favour genotypes of either parents or heterozygous alleles. Segregation distortion has been observed at morphological and molecular levels in the populations generated from heterozygotes derived from crosses between divergent parents in a wide range of organisms, such as in tomato (Rick, Reference Rick1966), maize (Helentjaris et al., Reference Helentjaris, Slocum, Wright, Schaefer and Nienhuis1986; Gardiner et al., Reference Gardiner, Coe, Melia-Hancock, Hoisington and Chao1993; Lu et al., Reference Lu, Romero-Severson and Bernardo2002), pearl millet (Liu et al., Reference Liu, Witcombe, Pittaway, Nash, Hash, Busso and Gale1994), wheat (Manabe et al., Reference Manabe, Ino, Kasaya, Takumi, Mori, Ohtsuka and Nakamura1999), sunflower (Tang et al., Reference Tang, Heesacker, Kishore, Fernandez, Sadik, Cole and Knapp2003), brassica (Saal & Struss, Reference Saal and Struss2005), moss (McDaniel et al., Reference McDaniel, Willis and Shaw2007) and Arabidopsis (Liu & Qu, Reference Liu and Qu2008).
In rice (Oryza sativa L.), segregation distortion is also commonly detected in the populations descended from hybrids derived from crosses involving divergent genotypes (McCouch et al., Reference McCouch, Kochert, Yu, Wang, Khush, Coffman and Tanksley1988; Sano, Reference Sano1990; Cheng et al., Reference Cheng, Saito, Takano and Ukai1996; Xu et al., Reference Xu, Zhu, Xiao, Huang and McCouch1997; Tan et al., Reference Tan, Vanavichit, Amornsilpa and Trangoonrung1998; Liu et al., Reference Liu, Zhu, Sun and Chen2001). The fundamental divergence in rice is divergence between indica and japonica subspecies (Chang, Reference Chang1976), which is a result of ecogeographical differentiation associated with genetic adaptability to environmental conditions. Indica rice adapts to tropical and subtropical environment at low latitude and altitude areas, whereas japonica rice adapts to temperate environment at higher latitude or higher altitude areas (Wang et al., Reference Wang, Second and Tanksley1992; Li & Rutger, Reference Li and Rutger2000; Zeng et al., Reference Zeng, Li, Yang, Shen, Zhang and Wang2001).
Analysis of segregation distortion based on F2 populations derived from reciprocal F1 hybrids grown at different altitudes will benefit to clarification of rice divergence caused by the effects of cytoplasm and altitude variation. In the present study, segregation distortion was investigated on morphological traits and molecular markers in six F2 populations generated from hybrid zygotes grown at three locations varied at altitudes from 400 to 2200 m. The results indicated that, besides nuclear genetic factors, indica cytoplasmic background and cytoplasmic male sterility (CMS) strongly affected segregation distortion, and altitude variation also had effects on segregation distribution. Besides segregation distortion, the six F2 populations were divergent into two groups according to their cytoplasm backgrounds.
2. Materials and methods
(i) Plant materials
Genetic stocks include two O. sativa cultivars, cv. XMG and cv. N34, F1 hybrids derived from reciprocal crosses between XMG and N34 and six F2 populations generated from the reciprocal F1 hybrids grown at three locations, which are less than 200 km apart in direct distance, but varied at altitudes from 400 to 2200 m (Table 1). XMG is a japonica landrace traditionally grown in fields at 2650 m altitude; while N34, a restorer used in japonica hybrid rice, is an inbred line developed by multiple crosses involving indica and japonica cultivars, and it possesses cytoplasm donated by indica cultivar IR8 (Hong et al., Reference Hong, Li, Wang, Shi, Huang and Tan2004; Tan et al., Reference Tan, Tan, Zhao, Zhang, Hong, Jin, Liu and Huang2004).
Table 1. Altitude and temperature of the three experiment locations

(ii) Collection and analysis of morphological data
Spikelet fertility (filled grains/total grains) and pollen fertility (fertile pollens/total pollens) were investigated with ten plants on two parents, and F1 hybrids grown at three altitude sites. Anthers were collected from spikelets just before flowering, pollen grains were observed under a microscope after suspended in 1% (w/v) I2–KI solution. Pollens in normal spherical shape and dark blue colour were regarded as fertile, pollens in irregular shape or without dark blue colour as sterile. Around 300 pollen grains from each sample were checked.
The two parents, two F1s, and six F2 populations were grown at 1860 m altitude (KM) in 2008. Nine morphological traits, including heading days, plant height, flag-leaf length, flag-leaf width, effective panicles, panicle length, panicle node length, total grains per panicle and spikelet fertility, were investigated on each progeny of six F2 populations.
(iii) Genotyping by PCR markers
Total DNA was extracted from leaves of each sample with the Cetyltrimethylammonium Bromide (CTAB) method developed by Murray & Thompson (Reference Murray and Thompson1980). The F2 progenies were genotyped with 16 polymorphic PCR markers screened in previous research. PCR primers were synthesized by Takara Biotechnology (Dalian) Co., Ltd. according to the sequence by McCouch et al. (Reference McCouch, Teytelman, Xu, Lobos, Clare, Walton, Fu, Maghirang, Li, Xing, Zhang, Kono, Yano, Fjellstrom, DeClerck, Schneider, Cartinhour, Ware and Stein2002), Komori et al. (Reference Komori, Ohta, Murai, Takakura, Kuraya, Suzuki, Hiei, Imaseki and Nitta2004) and Jiang et al. (Reference Jiang, Lu, Zhou, Wu, Zhuang, Liu and Mei2006). Each PCR mixture had a final volume of 12·5 μl, containing 20–40 ng of genomic DNA, 0·3125 unit of Taq DNA polymerase (Takara Biotechnology), 100 μM each of dNTP mixture, 160 nM of primers and 1·25 μl PCR buffer (Mg2+ Plus). The PCR was performed on an Applied Biosystems 2720 Thermal Cycler Set with the following thermal program: initial denature at 94°C for 5 min, followed by 40 cycles of 94°C for 30 s, 55°C (adjusted according to annealing temperature of each primer) for 30 s, and 72°C for 30 s, with a final extension at 72°C for 7 min. The PCR products were separated by electrophoresis in 2·5% agarose gel stained with ethidium bromide and photographed on an ultraviolet light-transmitting transilluminator.
(iv) Analysis of F2 segregation
Normal distribution fitness of morphological traits of the F2 populations was evaluated using Shapiro and Wilk's test (Shapiro & Wilk, Reference Shapiro and Wilk1965) with 7.05 version of DPS software (Tang, Reference Tang2004; http://www.chinadps.net/index.htm). Segregation distortion of morphological traits was measured by skewness. PCR marker segregation ratios were tested by Chi-square using Microsoft Office Excel 2003. Genetic divergence among the F2 populations was evaluated based on morphological traits and PCR markers. Genetic identities (I) and genetic distances (D) among the F2 populations were calculated with Nei's method (Reference Nei1972) based on the data of PCR markers and ten graded morphological traits. Dendrogram and phylogenetic tree were constructed with the UPGMA method (Sokal & Michener, Reference Sokal and Michener1958) by using DPS 7.05, and PopGen32 (Yeh et al., Reference Yeh, Yang, Boyle, Ye and Mao1997) and TreeView (Page, Reference Page1996), respectively.
3. Results
(i) Effects of cytoplasm and altitude variation on fertility of reciprocal F1 hybrids
The reciprocal F1 hybrids had significant differences on pollen and spikelet fertility. The fertility of the F1s derived from XMG×N34 was significantly higher than that of the F1s derived from N34×XMG at three altitudes. Especially, the difference in pollen fertility between the reciprocal hybrids was much higher (Table 2). The results suggested that cytoplasm had strong effects on fertility, especially on pollen fertility. Besides cytoplasm, altitude also affected fertility of the F1s. Pollen fertility of the F1s grown at low altitude was higher than that at high altitude. Pollen fertility of the reciprocal F1 hybrids was as high as 92·20 and 50·19% at 400 m altitude, but it was only 46·98 and 28·62% at 2200 m altitude (Table 2). However, the F1 hybrids grown at different altitudes did not show statistical differences on spikelet fertility. The results indicated that altitude variation had significant effects on pollen fertility of the F1 hybrids, but the effect was less obvious on spikelet fertility. The results suggested that altitude variation had selection pressure on pollen genotype, and such fertility difference could lead to genotype selection of descendant progenies.
Table 2. Pollen and spikelet fertility of the reciprocal F1 hybrids at the three altitudes
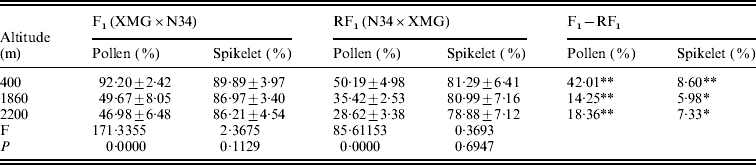
* and ** represent significant levels of P⩽0·05 and P⩽0·01, respectively. It is the same in the followings table.
(ii) Effects of cytoplasm and altitude variation on segregation distortion of morphological traits
Among nine morphological traits, only flag-leaf length fitted in the expected normal distribution in all six F2 populations (Fig. 1 i), the other eight traits were significantly (P<0·05 or P<0·01) deviated from normal distribution in almost all F2 populations. Three traits, effective panicles, panicle length and spikelet fertility, were significantly distorted toward N34 in all the populations (Fig. 1 a–c), while flag-leaf width and panicle node length were significantly skewed toward XMG (Fig. 1 d, e). All of those five traits showed severe distortion. The other three traits, total grains per panicle, plant height and heading days, were distorted toward different parents in different populations with low skewness values (Fig. 1 f–h).

Fig. 1. Skewness of morphological traits of the reciprocal F2 populations generated at the three altitudes. Skewness values of the F2 populations are represented by column height, in which black and line ones represent the populations with cytoplasm from XMG and N34, respectively. The ‘ns’ indicates the distortion is not statistically significant.
The F2 populations generated at different altitudes showed much different segregation distortions. Flag-leaf width had the largest skewness towards XMG in the two populations generated at 400 m altitude, while it showed smaller distortion in the populations generated at other locations (Fig. 1 d). Heading days showed the largest skewness in the populations generated at 2200 m altitude (Fig. 1 h), and panicle length and panicle node length showed the largest distortion in the populations generated at 1860 m altitude (Fig. 1 b, e).
Different traits showed different segregation distortions on different cytoplasms. For example, skewness values of spikelet fertility were larger in the populations with XMG cytoplasm than those in the populations with N34 cytoplasm (Fig. 1 c), while skewness values of panicle node length were larger in the populations with N34 cytoplasm (Fig. 1 e).
Besides, segregation distributions were also affected by interaction effects of cytoplasm and altitude variation. For examples, skewness value of panicle length was larger in the population with XMG cytoplasm generated at 1860 m altitude, but they were larger in the populations with N34 cytoplasm at other altitudes (Fig. 1 b). Interaction effects of cytoplasm and altitude were also observed on other traits, such as effective panicle (Fig. 1 a).
(iii) Segregation distortion of DNA markers
Segregation distortion of the F2 populations was also detected by DNA markers. Out of 16 loci, only six markers were in the Mendelian ratios, and 10 markers distributed on 7 chromosomes were significantly distorted from the expected 1:2:1 or 3:1 ratio (P<0·05 or P<0·01). Among them, six markers, which were located on chromosomes 2, 8, 9, 10 and 11, skewed toward N34, and three markers, which were located on chromosomes 3 and 5, skewed toward XMG; only RM218, which was located on chromosome 3, skewed favouring heterozygote (Table 3). Seven markers showed distortion in the populations generated at 400 m altitude; four and five markers showed distortion in the populations generated at 1860 and 2200 m altitudes, respectively.
Table 3. Marker segregation ratios of the F2 populations
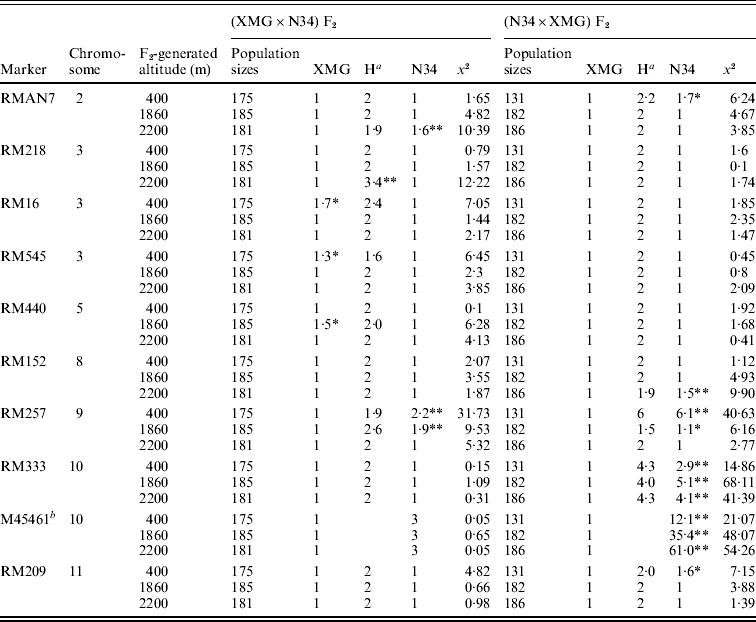
a Heterozygote.
b A dominant marker.
Only RMAN7 and RM257, which were located on chromosomes 2 and 9, respectively, were distorted in both of the reciprocal F2 populations, favouring N34, while most of markers were distorted just in one of the reciprocal populations, possessing cytoplasm of N34 or XMG. Furthermore, all the markers that showed distortion in the populations with N34 cytoplasm were skewed toward N34, while the markers that were distorted in the populations possessing XMG cytoplasm did not show the same favouring type, three favouring XMG, two favouring N34, one favouring heterozygote. The results indicated that cytoplasm from N34 had stronger effects than that from XMG on the segregation, especially for loci of RM333 and M45461. Marker M45461, a dominant marker (Fig. 2) located on chromosome 10, was extremely distorted towards N34 in the populations possessing N34 cytoplasm generated at the three altitudes, but it fitted in Mendelian ratio in the populations possessing XMG cytoplasm. Marker RM333 showed a similar segregation model (Table 3).

Fig. 2. Segregation of dominant marker M45461 in the reciprocal F2 populations generated at 1860 m altitude.
(iv) Divergence of the F2 populations
Both dendrogram on morphological traits and phylogenetic trees on PCR markers showed that the F2 populations were divergent according to the cytoplasm and the altitude variation. The six F2 populations were divergent into two groups (Fig. 3). One group consisted of the three populations generated from the same hybrids of XMG×N34 grown at different altitudes, which possessed cytoplasm from XMG. Another group consisted of the three populations derived from the reciprocal cross, which possessed cytoplasm donated by N34. In both groups, the genetic divergences between the populations generated at high altitude (2200 m) and low altitude (400 m) were larger than those between middle altitude (1860 m) and high altitude or between middle altitude and low altitude.

Fig. 3. Dendrogram (a) and phylogenetic tree (b) of the six F2 populations based on the Nei's genetic distances on morphological traits and molecular markers, respectively.
4. Discussion
Segregation distortion, which is a common case in segregated populations derived from crosses between divergent parents, can be caused by a number of physiological or nuclear genetic factors (Matsushita et al., Reference Matsushita, Iseki, Fukuta, Araki, Kobayashi, Osaki and Yamagishi2003; Liu et al., Reference Liu, Bernhardt, Jia, Wamishe and Jia2008). This study found that the effects of nuclear genes, cytoplasmic genes and altitude variation could cause segregation distortion. For instance, distortion of markers RMAN7 and RM257 was caused by nuclear genetic factors, since these two markers were distorted in both of the reciprocal populations, regardless of cytoplasm, especially for RM257, which was distorted in both of the reciprocal populations generated at the same altitudes. Nuclear genetic factors involved in segregation distortion, such as gametophytic selection gene, were also reported in other studies (Wilson and Levin, Reference Wilson and Levin1986; Matsushita et al., Reference Matsushita, Iseki, Fukuta, Araki, Kobayashi, Osaki and Yamagishi2003; McDaniel et al., Reference McDaniel, Willis and Shaw2007).
Although nuclear factors played a key role for segregation distortion, which was reported in a lot of studies (Bradshaw et al., Reference Bradshaw, Otto, Frewen, McKay and Schemske1998; He et al., Reference He, Li, Zheng, Shen, Lu, Chen and Zhu2001; Goloenko et al., Reference Goloenko, Davydenko and Shimkevich2002; Jiang et al., Reference Jiang, Lu, Zhou, Wu, Zhuang, Liu and Mei2006; Liang et al., Reference Liang, Peng, Ye, Li, Sun, Ma and Li2007; Liu et al., Reference Liu, Bernhardt, Jia, Wamishe and Jia2008), results of this study indicated that cytoplasm and altitude variation also had effects on distortion of morphological traits, although effects of dominant genes could not be excluded. Segregation distortions related to effects of cytoplasm were also reported in other studies (Faris et al., Reference Faris, Laddomada and Gill1998; Manabe et al., Reference Manabe, Ino, Kasaya, Takumi, Mori, Ohtsuka and Nakamura1999; Werlemark et al., Reference Werlemark, Uggla and Nybom1999; Goloenko et al., Reference Goloenko, Davydenko and Shimkevich2002).
Among the ten markers that showed segregation distortion, marker M45461, which was located within fertility restoring gene Rf-1 locus (Komori et al., Reference Komori, Ohta, Murai, Takakura, Kuraya, Suzuki, Hiei, Imaseki and Nitta2004), showed extreme distortion in the F2 populations possessing N34 cytoplasm, but its segregation fitted Mendelian ratio in the reciprocal populations possessing XMG cytoplasm. A reasonable cause for that is N34 possesses a fertility restoring gene, Rf-d1 gene, which is allelic to Rf-1 (Hong et al., Reference Hong, Li, Wang, Shi, Huang and Tan2004; Tan et al., Reference Tan, Tan, Zhao, Zhang, Hong, Jin, Liu and Huang2004) and CMS (Wang et al., Reference Wang, Zhang, Tan, Wen and Li2009), but XMG possesses normal cytoplasm. Therefore, it is expected that segregation of M45461 favoured N34 in the F2 populations on CMS cytoplasm. By contrast, it is also expected that segregation of this marker in the reciprocal F2 populations fitting Mendelian ratio, since the progenies did not possess CMS. Marker RM333, which is about 12·8 cM from M45461 (http://blast.ncbi.nlm.nih.gov/Blast.cgi and http://www.gramene.org), also showed similar distortion. Severe segregation distortion of these two markers could be caused by male gamete selection favouring the male gametes possessing Rf genotype due to interaction between CMS and Rf gene. Significant difference on pollen fertility between the reciprocal F1 hybrids was an evidence of the male gamete selection due to existence of CMS. Segregation differences of these two markers between the reciprocal F2 populations were a good example that interaction between cytoplasm and nuclear gene caused segregation distortion due to male gamete selection.
Besides CMS, cytoplasmic background also affected segregation distortion as reported in other studies (Faris et al., Reference Faris, Laddomada and Gill1998; Manabe et al., Reference Manabe, Ino, Kasaya, Takumi, Mori, Ohtsuka and Nakamura1999; Werlemark et al., Reference Werlemark, Uggla and Nybom1999; Goloenko et al., Reference Goloenko, Davydenko and Shimkevich2002). This study found that segregation distortions of the markers did not favour a special parent in the F2 populations possessing cytoplasm from XMG, but the distortions in the F2 populations possessing N34 cytoplasm favoured cytoplasm donor N34. A difference between the two cytoplasms is that cytoplasm possessed by N34 was from an indica cultivar through multiple crosses involved in indica and japonica cultivars (Hong et al., Reference Hong, Li, Wang, Shi, Huang and Tan2004), while the cytoplasm possessed by XMG is from a japonica landrace grown at fields with 2650 m altitude. The results are consistent with observation that distortions in the segregated populations possess indica cytoplasm favoured indica parents (He et al., Reference He, Li, Zheng, Shen, Lu, Chen and Zhu2001; Peng et al., Reference Peng, Liang, Wang, Wu, Li, Deng and Li2006; Liang et al., Reference Liang, Peng, Ye, Li, Sun, Ma and Li2007). These results indicated that japonica cytoplasm did not cause distortion favouring a special parent, but indica cytoplasm resulted in distortion favouring a maternal parent. The results also suggested that indica cytoplasm was not well compatible with japonica nuclear background, but japonica cytoplasm did not have such a problem with indica nuclei. According to such clues, japonica cultivars should be used as maternal parents in crosses between indica and japonica cultivars for developing japonica varieties.
Besides cytoplasm sources, results of this study also revealed that altitude variation had effects on segregation distortion. Segregation differences between the populations generated at different altitudes could be caused by the temperature differences among the three locations, which is an obvious difference of the environmental conditions among the locations. Segregation ratio affected by environmental conditions was also reported in Phytophthora sojae (MacGregor et al., Reference MacGregor, Bhattacharyya, Tyler, Bhat, Schmitthenner and Gijzen2002), and temperature stress resulting in segregation distortions were observed in other organisms, such as in Drosophila melanogaster (Mange, Reference Mange1968; Hiraizumi, Reference Hiraizumi1993) and tomato (Zamir et al., Reference Zamir, Tanksley and Jones1982).
This study revealed that the six populations were divergent into two groups on two cytoplasms, and then the groups were further differentiated according to similarity of altitudes where the populations were generated. The divergence could be resulted by difference of distortions among the populations, since distortion is a reflection of divergence (Tanksley & Nelson, Reference Tanksley and Nelson1996). The results indicated that cytoplasm and altitude had effects on divergent selection, which was a major factor for crop evolution (Michelmore & Meyers, Reference Michelmore and Meyers1998), but cytoplasm had much stronger effects than altitude factor.
This work was supported by National Natural Science Foundation of China (No. 30860065) and by Yunnan Provincial Science and Technology Department, China (2006NG05).








