We regret that the computer program used to generate the simulations of the examples presented in Owusu-Ansah, Curnow and Adu-Ampomah, Reference Owusu-Ansah, Curnow and Adu-Ampomah2013 contained an error. The corrected simulations for trial M2 now show that a single replication of many clones gives slightly higher selection gains than greater replication of fewer clones. The very small advantages of large numbers of years of harvest remain correct. The selection gains from the trials were considerably underestimated and, with the corrected simulations, the expected performance in terms of yield of the selected clones with the right combination of numbers of clones and replicates often exceeded the performance of the best standard clone. The selected clones still underperformed compared with the best standard clone when selection was based on percentage of diseased pods or numbers of healthy pods.
The Figures 1, 2 and 3 for selection based on yield, percentage diseased pods and numbers of healthy pods respectively replace the corresponding Figures in the original paper.
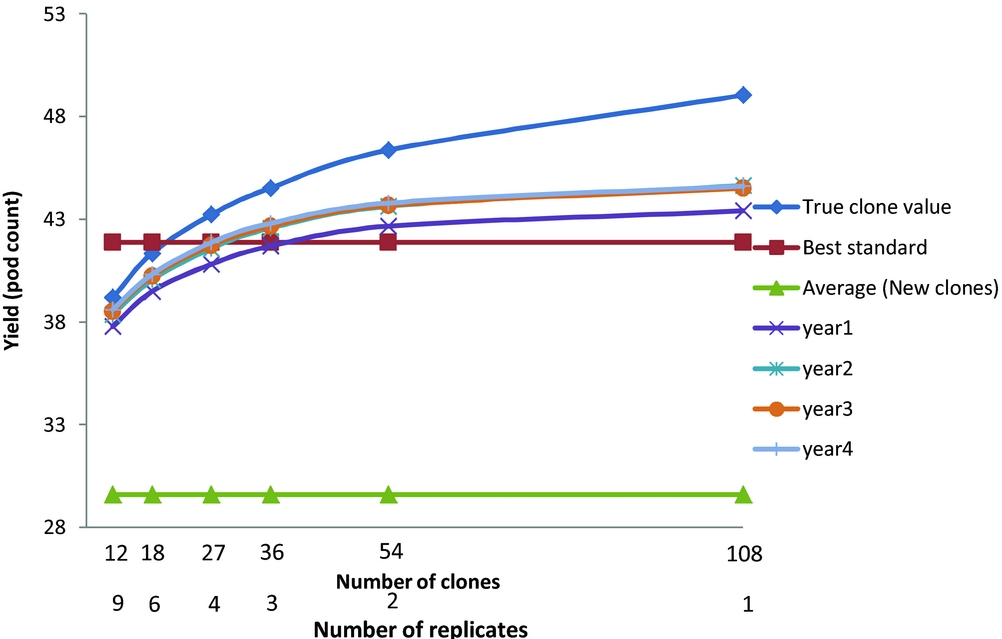
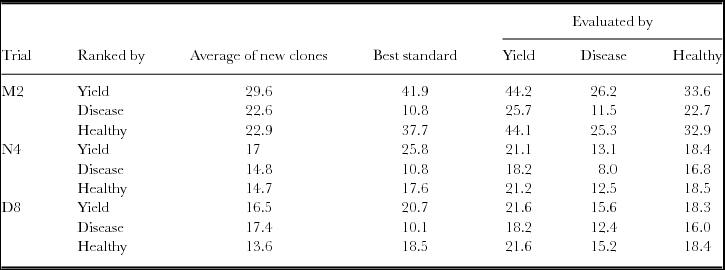
Figure 1. The yield response of the best three clones ranked by yield for different number of years and different numbers of clones compared to the true clonal values, best standard and average of new clones.
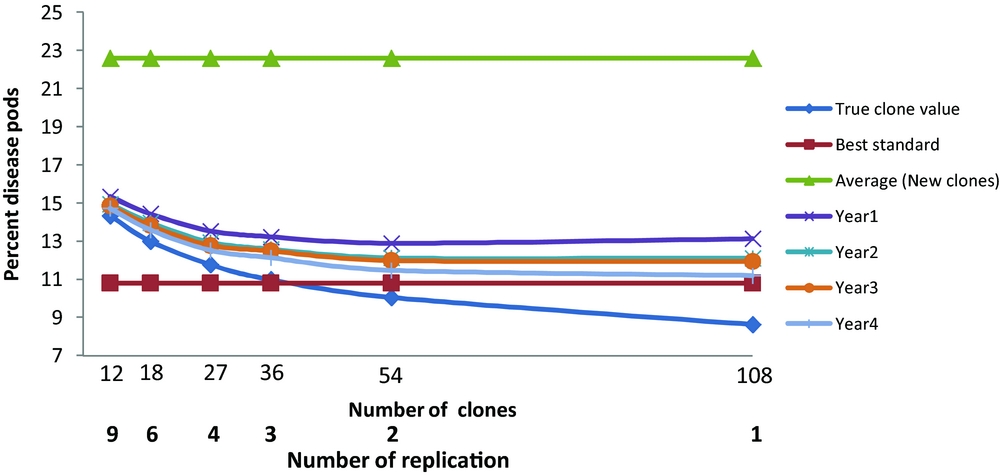
Figure 2. The percent diseased pods response of the best three clones ranked by percent diseased pods for different number of years and different numbers of clones compared to the true clonal values, best standard and average of new clones.
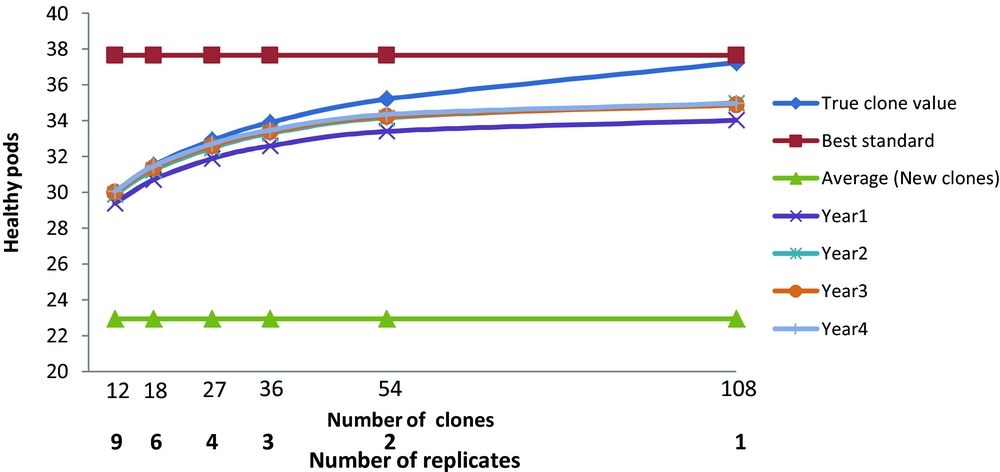
Figure 3. The healthy pods response of the best three clones ranked by healthy pods for different number of years and different numbers of clones compared to the true clonal values, best standard and average of new clones.
The actual trial, M2, had 18 clones grown in six replicate plots. This allows the comparison of differing numbers of new clones, N, and replicates, b, where Nb=108. Correction of the error resulted, when selection is based on yield (Figure 1) in the average yield of the best three new clones now exceeding the yield of the best standard when more than 36 new clones were tested. There were also advantages in basing selection on more than one year of harvest. The maximum expected gains were for a single replicate of 108 clones. The advantage of only one replicate over two was marginal and we recommend at least two replicates so that variance components can be estimated and the risk of missing values for any one clone reduced. The general conclusions with ranking and evaluation based on percentage diseased or on numbers of healthy pods were less affected with the best standard always outperforming the average of the best three selected clones; the expected gains were again maximised by having one replicate of 108 clones, and there were advantages in having data from more than one year of harvest.
Table 4 presented the results for M2 and two other trials, N4 and D8, showing the effects of selection by three different traits on gains evaluated in terms of these traits. Table 1 is the corrected version of Table 4 and is based on two replicates of 54 clones and four years of harvest. The general conclusions for M2 and D8 are similar to those of the original Table except for the better performance of the selected clones compared with the best standard already mentioned. In trial N4 the best three clones performed less well than the best standard in terms of yield but better than the best standard in percentage diseased and healthy pods.
Table 1. Average performance of best three clones as compared to the average of new clones and best standards for total yield, percent disease and healthy pods