Introduction
Biological cells are fundamental building blocks of tissues and organs of living organisms.Reference Alberts, Johnson, Lewis, Raff, Roberts and Walter1 Over their lifetimes, cells are responsible for the development, programmed remodeling, and regeneration of tissues and organs. In addition, cells play crucial physiological roles, such as transport of oxygen and carbon dioxide, signal transduction, muscle contraction/relaxation, homeostasis, immune response, and metabolic activities. In addition, a number of chronic and malignant diseases result from a wide range of genetic abnormalities and the conversion of normal cells to cancer cells.
Therefore, cells have been studied extensively as biomarkers for early detection, therapeutic targets, and even as sources for regenerative medicine that can take current levels of diagnosis and disease treatment to the next level.Reference Maitland and Collins2–Reference Rando7 In these efforts, cells are typically isolated from tissues of interest and cultured in vitro. Alternatively, various pluripotent and multipotent stem cells are being studied to generate specific cell types for use both in understanding their diverse emergent behavior and for treating disease and tissue defects via their in vivo transplantation.Reference Cherry and Daley8–Reference Addis and Epstein10 Efforts to directly convert fibroblasts (main component of connective tissue) to neuronal cells (i.e., nerve cells) are also under way.Reference Najm, Lager, Zaremba, Wyatt, Caprariello, Factor, Karl, Maeda, Miller and Tesar11,Reference Xue, Ouyang, Huang, Zhou, Ouyang, Li, Wang, Wu, Wei, Bi, Jiang, Cai, Sun, Zhang, Zhang, Chen and Fu12 Additionally, these cells are actively being used to assemble in vitro platforms, such as “body-on-a chip” that can screen newly designed drug molecules, including recombinant proteins and genes.Reference Williamson, Singh, Fernekorn and Schober13,Reference Esch, King and Shuler14 From these studies, there is a growing consensus that the successful use of cells in fundamental studies and translational clinical treatments greatly depends on the ability to retain cell viability and to regulate cellular activities in a controllable and predictable manner.
It is well understood that the diverse activities of cells, including growth, gene expression, migration, differentiation, and even programmed death, are regulated by multifaceted extracellular microenvironmental factors and genetic influences (Figure 1). The characteristics of the microenvironment include various soluble biochemical factors, biochemical and biomechanical properties of the surrounding extracellular matrix, and intercellular adhesion.Reference Kalluri15–Reference Leckband and Prakasam22 These factors also influence cellular binding and uptake of drug molecules and subsequent therapeutic activities.Reference Kong and Mooney23,Reference Kong, Liu, Riddle, Matsumoto, Leach and Mooney24 Major goals in the regenerative medicine field have been to recapitulate the extracellular microenvironment in vitro for specific diagnostic applications. In addition, a major goal for in vivo applications to regenerate damaged or diseased tissue has been to recapitulate specific tissue functions in vivo by first developing proper in vitro cell culture and subsequent in vivo transplantation tools and biomolecular carriers.Reference Discher, Mooney and Zandstra21,Reference Schmidt, Rowley and Kong25–Reference Brown, Bartolak-Suki, Williams, Walker, Weaver and Wong27 Aligned with these efforts, various materials, either alone or integrated with other engineering devices such as microelectromechanical systems, are also being explored to analyze and control chemical, mechanical, and biological interactions inside and between cells.Reference Khademhosseini, Langer, Borenstein and Vacanti28–Reference Zorlutuna, Jeong, Kong and Bashir34

Figure 1. Cross-talk between a cell and its extracellular microenvironment including soluble ligands (e.g., growth factors, cytokines), extracellular matrix (ECM), and external mechanical force. Various soluble ligands in biological fluids and insoluble ligands of the ECM activate or deactivate cellular signaling responsible for force generation, secretion of enzymes (e.g., metalloproteinase to soften ECM and transglutaminase to stiffen ECM), and secretion of other soluble ligands.
Interactive biological materials
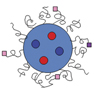
For the last few decades, there has been significant growth in designing such interactive biological materials by introducing or creating novel materials chemistry, characterization, and processing techniques. For instance, materials designed to present cell adhesion ligands found in the native extracellular matrix with mechanical properties similar to specific tissues were found to improve viability, modulate cellular adhesion machinery, and further stimulate or inhibit diverse cellular activities of various normal and pathologic cells.Reference Engler, Sen, Sweeney and Discher35–Reference Khetan, Guvendiren, Legant, Cohen, Chen and Burdick37 Additionally, various nanoscale materials (e.g., bioconjugated quantum dots, superparamagnetic iron oxide nanoparticles, carbon nanotubes) with unique photonic activities, as well as microfabricated materials coupled with electromechanical sensors were proposed to analyze cellular mechanics and signaling.Reference Sniadecki, Desai, Ruiz and Chen31,Reference Zrazhevskiy and Gao38–Reference Sniadecki, Anguelouch, Yang, Lamb, Liu, Kirschner, Liu, Reich and Chen43 To this end, nano-sized biomolecular carriers were developed to facilitate binding to target cells and subsequent intracellular drug transport (Figure 2).Reference Rajendran, Knoelker and Simons44–Reference Wong, McDonald, Taylor-Pinney, Spivak, Kaplan and Buehler47 However, there are still significant grand challenges in assembling such material systems that interface with biological cells. These challenges include overcoming both the difficulty of assembling multifunctional materials at varied length scales and the limited controllability of the feedback mechanism between cells and materials, and realizing the ability to sense and detect cellular near- to mid-term activities in vivo.

Figure 2. Multifunctional nanoparticles devised to image and treat target cells and tissues. Particles are loaded with image contrast agents and drugs or interests and their surfaces are engineered to present peptides or proteins that can specifically bind with proteins overexpressed by target cells and tissues. Note: MRI, magnetic resonance imaging; CT, computed tomography.
This issue of MRS Bulletin addresses these scientific and engineering challenges. The articles in this issue first critically explore the design of materials that have proved capable of reporting biological activities of cells, and second, identify challenges that are currently being faced. In addition, recent work that incorporates molecular and cellular biology techniques, specifically genetic engineering and strategies that harness the immune system into materials design, is described. The articles also include a discussion of opportunities for the future development of multifunctional materials for sensing and therapies. Finally, clinical perspectives and opportunities for the future development of these materials are discussed.
Overview of the articles
Kuo and Smith discuss perspectives on natural tissues as materials and also discuss how to develop novel synthetic materials that guide cell function. Next, Lewis et al. introduce efforts to design materials capable of modulating the immune response: (1) materials capable of suppressing immune response and (2) materials capable of activating or enhancing immune response. Then, Mastria and Chilkoti present a body of work based on genetically encoded elastin-like polypeptides that offer a route to modular, multifunctional drug delivery systems. Hwang et al. discuss the area of nanotheranostics and give examples of their recent work on genetic modification of cells and molecular imaging. Lee et al. introduce material considerations for vascularized organs-on-a chip, which is rapidly emerging as a promising tool for fundamental and applied biological studies. Finally, Gomes et al. present their perspective on the potential barriers for translating gene therapies into clinical practice and what must be overcome for clinical translation.
Glossary
Apoptosis: The process of programmed cell death.
Extracellular matrix: It is composed of proteins and other molecules (including polysaccharides) that surround cells.
Signal transduction: The cascade of processes by which an extracellular signal (typically a hormone or growth factor) interacts with a receptor at the cell surface, causing a change in the level of a second messenger and ultimately affects a change in the cell function.
Homeostasis: The tendency of an organism or a cell to regulate its internal conditions.
Pluripotent stem cell: A cell that possesses the potential to take on many fates in the body, including all of the more than 200 different cell types.
Multipotent stem cell: A cell that possesses the ability to differentiate into a various but limited number of cell types, especially into cells of a closely related family of cells.
Gene expression: The conversion of the information from the gene into mRNA (messenger RNA) via transcription and then to protein via translation.
Cell differentiation: The normal process by which a less specialized cell develops or matures to possess a more distinct form and function.