To the Editor:
In the article entitled ‘Molecular analysis of high-level ciprofloxacin resistance in Salmonella enterica serovar Typhi and S. Paratyphi A: need to expand the QRDR region? by Capoor and co-workers [Reference Capoor1], the authors have described new substitutions in DNA gyrase subunit, GyrA. DNA gyrase (topoisomerase II) is a multimeric enzyme composed of two pairs of subunits, Gyr A (878 residues) and GyrB, and the enzyme is inhibited by quinolones, like nalidixic acid, and fluoroquinolones, such as ciprofloxacin [Reference Drlica2]. Resistance to nalidixic acid is often mediated by substitutions in position Ser83 or Asp87 of GyrA while resistance to both nalidixic acid and ciprofloxacin frequently is the result of substitutions in both positions [Reference Hirose3, Reference Izumiya4]. The positions 83 and 87 are parts of the so-called quinolone resistance-determining region (QRDR) which has been defined by Yoshida and co-workers to cover the region between position 67 and 106 of GyrA [Reference Yoshida5], but Capoor and colleagues have proposed a highly relevant expansion of QRDR [Reference Capoor1].
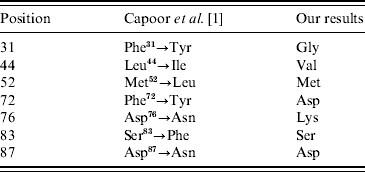
They have also described new amino-acid replacements in GyrA which are Phe31→Tyr, Leu44→Ile, Phe72→Tyr, Asp76→Asn, Glu133→Gly [Reference Capoor1] as well as previously described substitutions, i.e. Met52→Leu [Reference Capoor6], Ser83→Phe, and Asp87→Asn [Reference Ling7, Reference Eaves8]. The Leu44→Ile, Phe72→Tyr, Asp76→Asn substitutions have previously been described by Capoor and colleagues [Reference Capoor6]. Consensus is that e.g. Phe31→Tyr denotes a replacement of the predominant Phe residue in position 31 by a Tyr residue.
With a nucleotide sequence encompassing nt 76–480 from the gyrA gene of Salmonella Typhimurium strain 14028S [GenBank acc. no. CP001363] as query sequence we searched the nr/nt, refseq_genomic, chromosome, and wgs databases of the NCBI GenBank (http://www.ncbi.nlm.nih.gov/) for hits by Megablast with an expected threshold of 10−4. We also used Salmonella (taxid: 590) as the optional organism. Subsequently, we searched the nucleotide database by using the words ‘gyrA AND Salmonella’ and got additional hits.
In total, we achieved 358 entries (last accessed on 8 June 2010) all from different strains of divergent serovars of Salmonella. Owing to very low identity to GyrA or too short sequences only covering 11 positions including positions 83 and 87, 26 of the entries were discarded.
The resulting 332 sequences were analysed to determine the nature of the residues in positions 31, 44, 72, and 76 of GyrA. All 332 sequences contained the positions 72 and 76, but owing to paucity of sequence, 237 sequences also included position 44, whereas only 94 sequences also covered position 31. Ultimo 2007, 162 partial or complete sequences of GyrA from Salmonella have been released in the GenBank databases.
Each of the 332 sequences was carefully analysed, and our results showed that position 31 was inevitably occupied by Gly, residue 44 by Val, residue 72 by Asp, and position 76 by Lys (Fig. 1). These results are different from those reported by Capoor and co-workers [Reference Capoor1] as seen in Table 1. Amino-acid replacements were not found in any of these four positions among all the analysed Salmonella sequences.

Fig. 1. ClustalW alignment of a section including residues 29–89 of non-redundant genomic GyrA sequences from different serovars of S. enterica subsp. enterica and S. enterica subsp. arizonae serovar 62z4,z23 – in comparison with GyrA from Escherichia coli K-12 substrain MG1655. The residues discussed in the text are underlined. The GenBank accession nos. are: Typhi CT18, NC_003198; Typhi Ty2, NC_004631; Typhimurium, NC_003197; Dublin, NC_011205; Javiana, NZ_ABEH02000005; Kentucky, NZ_ABEI01000001; Tennessee, ACBF01000027; 62z4,z23–, NC_010067; Virchow, NZ_ABFH02000001; Gallinarum, NC_011274; Schwarzengrund, NC_011094; Typhi 2114, AM283471; Choleraesuis, NC_006905; E. coli K-12, NC_000913.
Table 1. Comparison of the amino-acid replacements in different positions of GyrA reported by Capoor et al. [Reference Capoor1] and the predominant residues in the same positions found by analyses of 332 GyrA sequences
Amino-acid substitutions in GyrA from Salmonella are occasionally related to the sequence of GyrA from Escherichia coli, therefore we also examined the equivalent sequences of GyrA from this species. Of the 163 E. coli available sequences in the GenBank databases, we found in all instances Gly31, Val44, Asp72, and Lys76.
The discrepancies between our results and those reported by Capoor and colleagues [Reference Capoor1, Reference Capoor6] could be caused by different numbering of the residues because the precursor GyrA protein may be processed into a signal peptide and a functional enzyme. However, this is contradicted by the numbering of the residues, Met52, Ser83, and Asp87, which is in accordance with an unprocessed precursor GyrA protein. That precursor GyrA remains unmodified has also been corroborated by the absence of predicted signal peptides and the prediction of the subcellular localization of GyrA. The web-based applications, SignalP v. 3.0 (http://www.cbs.dtu.dk/services/SignalP/) [Reference Bendtsen9] and PSORTb v. 3.0 (http://www.psort.org/) [Reference Gardy10] were used for prediction of putative signal peptide sequences and the subcellular localization of GyrA, respectively. Signal peptides were not predicted by either Neural Networks or Hidden Markov models, and the score for a predicted cytoplasmic location was 9·26 whereas the next highest score was 0·48 for a predicted periplasmic location. These predictions are consistent with the known function of GyrA which takes place in the cytoplasma. The precursor GyrA thus remains unprocessed.
Based on our analyses, it is our opinion that the positions of the amino-acid replacements, Phe31→Tyr, Leu44→Ile, Phe72→Tyr, and Asp76→Asn, described by Capoor and co-workers [Reference Capoor1] are debatable; to our knowledge, GenBank entries documenting the replacements characterized by Capoor et al. have not been released.
The positions of the described Met52→Leu, Ser83→Phe, and Asp87→Asn substitutions are compatible with the Salmonella GyrA sequences (see Fig. 1). The Ser83→Phe and Asp87→Asn substitutions have also been reported in several other publications [Reference Eaves8, Reference Bendtsen9].
We do not dispute the nature of described substitutions by Capoor et al. [Reference Capoor1], but we have justifiable reservations about their correct positions in the GyrA sequence. Our analyses of the available 332 GyrA sequences have unequivocally shown that Phe31, Leu44, Phe72, and Asp76 are not found in any of the GenBank sequences. If the positions are maintained it consequently means that Phe31, Leu44, Phe72, and Asp76 themselves are substitutions. Therefore, we consider the reported Phe31→Tyr, Leu44→Ile, Phe72→Tyr, and Asp76→Asn substitutions in GyrA as controversial and possibly incorrect.
In their article, Capoor et al. [Reference Capoor1] have corroborated the Phe72→Tyr substitution by referring to an Asp72→Gly substitution in Salmonella Senftenberg described by Eaves et al. [Reference Eaves8]. The latter substitution is consistent with a dominant Asp residue in position 72 of GyrA (Fig. 1).
Regarding the Glu133→Gly substitution [Reference Capoor1], it is correct that Glu is the prevalent residue in position 133 of GyrA among the different serovars of Salmonella, apart from Salmonella Typhi. Position 133 in GyrA is covered by 315 of the obtained GyrA sequences from GenBank, and 214 of these sequences are from serovars other than Typhi. However, only two sequences (GenBank acc. nos. AY302581 and AY302582) among the 214 sequences carry a Glu133→Gly substitution indicating that Glu133 is the dominant residue in GyrA from these serovars. In E. coli, Glu133 is also found in all sequences.
This picture is reversed in GyrA from S. Typhi because only four of the 101 sequences that cover position 133 hold a Glu133 residue (GenBank acc. nos. AY302585, AY302588, GU190905, EF176634) while the remaining 97 sequences contain Gly133.
These findings make it reasonable to assume that the genuine substitution in position 133 of GyrA from S. Typhi is in fact Gly133→Glu and not Glu133→Gly. Among the 162 Salmonella GyrA sequences available in the GenBank database Ultimo 2007, 90 sequences were from S. Typhi, and 87 of these contained a Gly133 residue while only three had a Glu133 residue.
Some of the amino-acid replacements reported by Capoor and colleagues [Reference Capoor1] form the basis of their conclusion on an expansion of QRDR, but owing to the dubious positions of some of the described substitutions as shown in this letter, this conclusion may need a reassessment.
Declaration of Interest
None.
The authors reply
We congratulate Drs Walther-Rasmussen and Høiby on their work. As regards our article, which was accepted for publication in 2008, our findings were the result of collaboration of a group from the National Institute of Communicable Diseases (NICD) in New Delhi and the Pasteur Institute in Lille.
It may be that the variation in our data may be due to regional differences in isolates. Our sequencing was undertaken during 2005 and 2006. We had hoped to be able to repeat the sequencing and retrieve the data. Unfortunately, this has not proved possible; the laboratory at the Pasteur Institute has closed and two co-authors from NICD have moved to other countries.




