INTRODUCTION
African countries have in recent years experienced more epidemics and cases of cholera than countries in Asia and America [1, 2]. After the seventh pandemic pathogen Vibrio cholerae O1 biotype El Tor reached and initially spread in West African countries in the 1970s, the majority of countries in the eastern and southern parts of Africa have experienced major cholera epidemics, occasionally with unusually high mortality rates. The most recent statistics show that in 2004 a total of 95 560 cases were reported from 31 countries of Africa which accounted for 94% of the global total of officially notified cholera cases [3]. A 23% increase in the number of reported deaths by cholera compared to 2003 was observed in 2004, with an overall case-fatality rate of 2·4% for the continent [3]. In contrast to the V. cholerae strains causing outbreaks in South-east Asia, which have been extensively characterized, little information is available about the characteristics of the recent epidemic strains implicated in cholera outbreaks in Africa [Reference Dalsgaard4].
The most recent epidemic in Guinea–Bissau began in October 1996, extended throughout 1997, and included a total of 26 967 reported cases. During the epidemic, the National Public Health Laboratory in Bissau, Guinea–Bissau, monitored levels of antimicrobial resistance and this showed the emergence of a multidrug-resistant strain during 1997, concomitant with the second peak of the epidemic. At the same time, an increase in the case-fatality rate was observed [Reference Dalsgaard5]. A huge outbreak occurred in Mozambique in August 1997 caused by V. cholerae O1 biotype El Tor with the number of cases totalling more than 10 000 by the end of 1997. The epidemic continued into 1998 and 1999, with V. cholerae O1 being introduced into South Africa in 1998 by migrant workers from Mozambique, in particular to the provinces of Gauteng and Mpumalanga, the latter bordering Mozambique [Reference Athan, Donohue and Durrheim6]. Although all the cases in Gauteng were identified in migrant labourers, many of those in Mpumalanga had acquired the infection in South Africa through contamination of local water sources [Reference Athan, Donohue and Durrheim6]. In these epidemics, the patients were treated with fluid and electrolyte replacement therapy; in addition, antibiotics were found to be very effective in shortening the duration of episodes and thus helped reduce the case fatality of cholera substantially. However, resistance to cotrimoxazole and tetracycline was reported. Subsequently, cases of cholera were identified in children in an informal settlement in Kwazulu-Natal in 1999, the third province to have been affected (communication from the Department of Health of South Africa). During 2004, major outbreaks of cholera occurred in Cameroon, Chad, Guinea, Mali, Niger, Senegal, and Zambia [3]. However, the characteristics of the V. cholerae O1 strains responsible for these outbreaks, including their genotypes and mechanisms of antibiotic resistance remain mostly unknown [Reference Dalsgaard4].
Although little is known about the antibiotic susceptibility of epidemic V. cholerae strains, particularly in African countries, it appears that antibiotic-resistant V. cholerae strains are increasing worldwide. Plasmids are known to encode and transfer resistance in V. cholerae O1 [Reference Glass7, Reference Weber8]. The SXT element, which has properties similar to those of a conjugative transposon, was also found to carry genes encoding resistance to trimethoprim–sulphamethoxazole (TMP–SMX) and streptomycin in V. cholerae O139 and O1 strains isolated in India, but was not present in O1 strains obtained in 1994 from Rwandan refugees in Goma, Zaire [Reference Waldor, Tschape and Mekalanos9]. Several studies implicated class 1 integrons with multiple antibiotic resistance and these have been described as vehicles for possible horizontal acquisition of resistance gene cassettes [Reference Jones10, Reference Dalsgaard11]. Zambian people have a long history of exposure to cholera and V. cholerae O1 and many cholera outbreaks have occurred there since 1971. However, the genotypic and phenotypic characteristics of V. cholerae causing these outbreaks remain unknown. This study was initiated to obtain a clear picture of the patterns of antibiotic resistance and gain an understanding of the genetic traits of epidemic strains of V. cholerae O1 isolated in Zambia during the outbreaks that occurred between 1990 and 2004.
MATERIAL AND METHODS
Outbreak data
V. cholerae O1 strains isolated from eight outbreaks, between the period of February 1990 and December 2004, were tested for antimicrobial resistance. Antibiotic resistance was monitored depending on the availability of antibiotic discs at that given time. Data on the cholera outbreaks and V. cholerae O1 isolates were compiled from laboratory records.
Bacterial isolates
Sixty-nine V. cholerae isolates, isolated between 1996 and 2004, were examined. They were recovered from stool or rectal swabs of suspected cholera patients from different cholera treatment centres and the University Teaching Hospital in Lusaka. Isolation and identification were performed according to WHO standards [12]. Specimens were cultured on thiosulphate–citrate–bile salts–sucrose agar (Eiken Ltd, Tokyo, Japan) before and after pre-enrichment in alkaline peptone water (pH 8·6) and on blood agar. Colonies with the characteristic appearance of V. cholerae were confirmed by biochemical and serological tests using polyvalent O1 and mono-specific Ogawa and Inaba antisera (Denka Seiken, Tokyo, Japan).
Susceptibility testing
Susceptibility to antimicrobial agents was determined by the Kirby–Bauer disk diffusion method and interpreted as recommended by the National Committee for Clinical Laboratory Standards [13, Reference Garg14] with commercial antimicrobial discs (Oxoid, Basingstoke, UK): ampicillin (10 μg), TMP–SMX (25 μg), cefotaxime (30 μg), tetracycline (30 μg), norfloxacin (10 μg), furazolidone (100 μg), erythromycin (15 μg), doxycycline (30 μg), and chloramphenicol (30 μg). Escherichia coli ATCC 25922 strain susceptible to all antibiotics tested was used as a control.
Plasmid analysis
Plasmid DNA was prepared by the rapid alkaline lysis method of Kado & Liu [Reference Kado and Liu15] with some modifications [Reference Talukder16]. Plasmid DNA was separated by horizontal electrophoresis in 0·7% agarose (Sigma Chemical Co., St. Louis, MO, USA) slab gels in Tris–borate EDTA (TBE) buffer (Bio-Rad, Hercules, CA, USA ) at room temperature at 100 V (50 mA) for 3 h. The gel was stained with ethidium bromide and video images were recorded on a gel documentation system. Plasmid DNA was sized against reference plasmids in E. coli strain PDK-9 [Reference Haider17].
A nalidixic acid-resistant mutant strain of E. coli K-12, strain J53-1 (lac + pro met Nalr) was used as the recipient in conjugation experiments. After mating on non-selective L agar (Difco, Detroit, MI, USA) incubated at 37°C for 4–6 h, transconjugants were harvested and appropriate dilutions were spread on plates of Fluorocult E. coli O157:H7 agar (Merck, Virum, Denmark) supplemented with 40 mg/l nalidixic acid and adequate concentrations of selected drugs; 10 mg/l tetracycline, 25 mg/l streptomycin and 32 mg/l trimethoprim were used for selection together with 40 mg/l nalidixic acid. Phenotypic appearance was used to differentiate possible spontaneous nalidixic acid-resistant donors from transconjugants which were screened for the presence of the resistance plasmid.
Molecular analysis
The 69 strains of V. cholerae O1 were examined using a PCR-based method for the presence of ctxA (a gene encoding the B subunit of cholera toxin), tcpA (encodes for biosynthesis of the toxin co-regulated pili), rstR (encodes for the repressor gene for the CTX prophage), SXT element (encodes the resistance markers for streptomycin and trimethoprim [Reference Dalsgaard4], and intl1 (encodes class 1 integrons). [Reference Dalsgaard4] Oligonucleotide primers used in the PCR assays, their sequences and the amplicon sizes are shown in Table 1. The strains were stored in LB broth with 25% glycerol at −70°C and subcultured on LB agar. A single colony was confirmed by serology using appropriate polyclonal antisera, inoculated in 3 ml LB broth and incubated at 37°C in a shaking water bath at 120 rpm for 18 h. One ml of broth culture was transferred to an Eppendorf tube and centrifuged at 6000 g for 6 min. The pellet was resuspended in 1 ml sterile deionized water, boiled for 10 min and held on ice for 20 min. The samples were centrifuged at 12 000 g for 10 min and the supernatant was used as the template DNA for PCR.
Table 1. Oligonucleotide primers, sequences, and amplicons used in PCR assaysFootnote *

* Primers were for the detection of pathogenic and antibiotic resistance-related genes in V. cholerae O1 strains isolated during cholera outbreaks between 1996 and 2004 in Zambia.
The following were added to 22 μl PCR mixture: 2·5 μl Mg2+-free 10× amplification buffer [500 mm KCl, 200 mm Tris–HCl (pH 8·4)], 0·75 μl of 50 mm MgCl2, 2·5 μl each of 2·5 mm dATP, dTTP, dGTP, and dCTP, 7·5 pmol each of the primers as described before [Reference Nusrin18–Reference Burrus and Waldor21], and 1 U Taq DNA polymerase (Invitrogen, Life Technologies, Carlsbad, CA, USA), 14·55 μl sterile deionized water. PCR was carried out in 0·2 ml PCR tubes with 22 μl PCR mixture and 3 μl template DNA. PCR was performed in an automated thermocycler (PTC-0200 Peltier Thermal Cycler, MJ Research Inc., Watertown, MA, USA). The amplification conditions were set at one cycle of 94°C for 5 min, followed by 35 cycles at 94°C for 40 s, 50°C for 40 s, and 72°C for 90 s with a final extension of 72°C for 7 min. The PCR products were separated by electrophoresis in 1% agarose gel and visualized under UV light following staining with ethidium bromide. Three strains 154 V. cholerae O1 (classical), 196318 V. cholerae O139 and SCE-188 V. cholerae non-O1/non-O139 (environmental) were used as standard reference strains. The sequences of the primers used in this study are identical to those described previously [Reference Nusrin18–Reference Burrus and Waldor21].
RESULTS AND DISCUSSION
The antibiotic resistance data, made available from laboratory records during epidemics between 1990 and 2004, on V. cholerae serogroup O1 isolated during eight cholera outbreaks, are shown in Table 2. A low level of resistance (2–3%) to tetracycline was recorded in the first two cholera outbreaks that occurred during 1990–1991. However, due to continued use for therapy and prophylaxis, resistance increased dramatically to tetracycline (95%) along with chloramphenicol (78%), doxycycline (70%) and TMP–SMX (97%) in subsequent outbreaks in 1992. Interestingly, the adoption of a national policy to replace tetracycline with erythromycin in treating cholera, led to a significant drop in resistance to tetracycline and chloramphenicol during 1993–2002. It is unknown whether the isolates responsible for earlier and recent epidemics are of the same clonal origin. Again, a low level of erythromycin resistance (2%) was observed during the outbreaks that occurred between 1998 and 2002, but a steady increase in the pattern of resistance to chloramphenicol and complete resistance to cotrimoxazole (100%) continued until 2004. The association between the development of resistance to tetracycline, chloramphenicol, and cotrimoxazole with large-scale use of antibiotics for treatment and prophylaxis of cholera is well recognized [Reference Glass22–Reference Finch24]. Our surveillance data showed that multi-drug resistant (MDR) strains of V. cholerae O1 emerged during these outbreaks presumably due to heavy and widespread use of antibiotics for therapy. The MDR strains were resistant to ampicillin, tetracycline, and TMP–SMX, which was alarming.
Table 2. Antibiotic resistance of V. cholerae O1 isolated between February 1990 and December 2004 from cholera outbreaks in Zambia
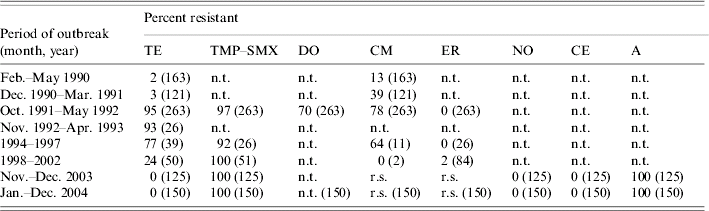
r.s., Reduced susceptibility; n.t., not tested.
TE, Tetracycline; TMP–SMX, trimethoprim–sulphamethoxazole; DO, doxycycline; CM, chloramphenicol; ER, erythromycin; NO, norfloxacin; CE, cefotaxime; A, ampicillin.
Numbers in parentheses indicate number of strains examined.
Table 3 shows that V. cholerae O1 strains from 1996/1997 were resistant to TMP–SMX and tetracycline whereas those strains from 2003/2004 were resistant to TMP–SMX and furazolidone, but susceptible to tetracycline. The emergence of resistance to various antibiotics amongst vibrios is a well established phenomenon [Reference Dalsgaard4, Reference Glass7, Reference Garg14]. For management of cholera, tetracycline is used as the first-line drug [Reference Dalsgaard4] but due to the emergence of resistance it has been replaced with erythromycin in many cases. The antibiotic of choice must always be based on an updated culture and sensitivity report. Since the emergence of such resistance amongst V. cholerae against antibiotics may significantly influence the future strategies of controlling cholera, continuous monitoring of the epidemic strains is thus crucial.
Table 3. Antibiogram, plasmid analysis and genetic screening of V. cholerae O1 strains isolated from Zambia
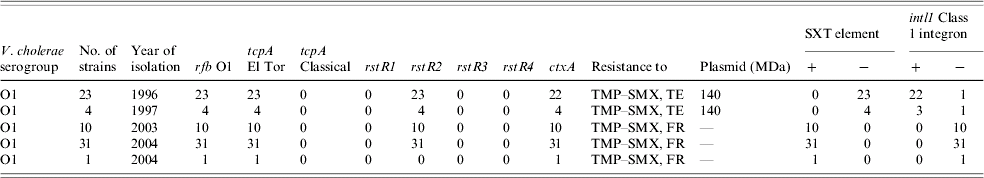
TMP–SMX, Trimethoprim–sulphamethoxazole; TE, tetracycline; FR, furazolidone.
Plasmid analysis revealed the presence of a conjugative 140 MDa MDR plasmid among strains from 1996 to 1997 (Table 3). These plasmid-carrying strains were resistant to TMP–SMX and tetracycline. On the other hand, TMP–SMX-resistant but tetracycline-susceptible strains isolated during 2003–2004 did not harbour any plasmids indicating plasmid-mediated resistance to tetracycline. Our findings appear to be in agreement with those of Waldor et al. [Reference Waldor, Tschape and Mekalanos9] who also confirmed that the SXT element and its resistance genes to sulphonamides, trimethoprim and streptomycin were transferable. However, the lack of plasmids in 2003–2004 strains suggests that the resistance determinants in these MDR strains were not transferable. A resistance gene cassette (aadA2) was found in chromosomally located class 1 integrons in the studies of V. cholerae O1 isolates from Vietnam and O1 isolates from Thailand [Reference Dalsgaard11], Italy, and India [Reference Dalsgaard4, Reference Shi20]. In the Indian strains, resistance to TMP–SMX and streptomycin in some cases was found to be due to the SXT element [Reference Waldor, Tschape and Mekalanos9]. Results of our PCR assays show that all but one strain harbouring the conjugative 140 MDa MDR plasmid were positive for intl1 class 1 integron but all lacked the SXT element (Table 3). Conversely, all 42 strains lacking this plasmid gained resistance to furazolidone but lost the tetracycline resistance. Unlike the tetracycline-resistant strains from 1996/1997, all tetracycline susceptible but furazolidone-resistant strains from 2003/2004 contained the sxt element but not intl1 class 1 integron. This study thus shows clearly that the tetracycline-resistance determinants in the MDR strains from 1996/1997, were encoded in the plasmid-borne class 1 integron [Reference Jonson, Holmgren and Svennerholm25] and that the furazolidone resistance in MDR strains of 2003/2004 were encoded in the SXT element [Reference Dalsgaard4] but not in the class 1 integron.
Unlike most studies that have reported the resistance determinants in MDR strains to be encoded in the class 1 integrons [Reference Jonson, Holmgren and Svennerholm25], SXT element [Reference Dalsgaard4], or transposons [Reference Waldor, Tschape and Mekalanos9], the resistance determinants for TMP–SMX in the present study were encoded neither in the SXT element nor in the class 1 integrons or plasmid. This might suggest integration of this resistance gene in the host genome and the same assumption may be true in the case of a tetracycline-resistant strain from 1996/1997 which was negative for the class 1 integron. In any case, the spread of antibiotic resistance in microbes has been attributed to the mobilization of drug resistance markers by a variety of agents such as plasmids, transposons, and integrons [Reference Dalsgaard4, Reference Waldor, Tschape and Mekalanos9, Reference Dalsgaard11, Reference Tabtieng23].
All 69 strains were serologically confirmed as serogroup O1 biotype El Tor of V. cholerae. Genetic screening by PCR revealed that all, except two, one isolated during 1997 lacking ctxA and the other isolated during 2004 lacking rstR2, had important epidemic markers namely rfbO1, ctxA, rstR2, and tcpA El Tor. We used the repressor gene rstR as a genetic biotype marker since recent studies in Mozambique have shown that V. cholerae O1 strains associated with cholera outbreaks are a variant of El Tor biotype in that they carry the classical CTX prophage [Reference Ansaruzzaman26]. Such kind of hybrids strains of V. cholerae O1 were previously reported in Bangladesh [Reference Nusrin18, Reference Nair19]. Since Zambia shares a border with Mozambique, the rationale for the molecular analysis was to understand if such hybrid El Tor strains had spread to Zambia. This study shows that the Zambian V. cholerae El Tor strains are not hybrids and are typical seventh pandemic strains of the El Tor biotype. The strain that was negative for ctxA but positive for El Tor tcpA and rstR2 of the cholera toxin gene clusters may be a non-toxigenic V. cholerae O1, which is not new because such strains have previously been reported from diarrhoea and other extra-intestinal infections [Reference Jonson, Holmgren and Svennerholm25]. Accordingly, the strain that was ctx positive but rstR negative may represent a genotypic variant. Although we have not examined the strains further for ctxB or other neighbouring genes of the CTX prophage gene clusters, we speculate that these strains may have a truncation in CTX prophage region.
ACKNOWLEDGEMENTS
The core donors to the International Centre for Diarrhoeal Disease Research, Bangladesh (ICDDR,B) supported this work. Current donors providing unrestricted support include the aid agencies of the Governments of Australia, Bangladesh, Belgium, Canada, Japan, Kingdom of Saudi Arabia, The Netherlands, Sweden, Sri Lanka, Switzerland, and the United States of America.
DECLARATION OF INTEREST
None.





