INTRODUCTION
Extraintestinal pathogenic Escherichia coli (ExPEC), cause a wide spectrum of illnesses including cystitis, pyelonephritis, bacteraemia, prostatitis and other infections which occur outside the human intestine. The most common type of infection due to ExPEC is urinary tract infection (UTI); 70–95% of UTIs are caused by ExPEC [Reference Hooton and Stamm1–Reference Stamm and Hooten3]. It is estimated that 11% of women aged ⩾18 years are affected by UTIs annually, resulting in over 1 billion dollars of direct and indirect costs per year [Reference Russo and Johnson2, Reference Foxman4, Reference Foxman5]. The increasing antibiotic resistance of the E. coli that commonly cause UTIs has complicated their management.
ExPEC colonize the human intestine and then are transferred to an extraintestinal site, such as the bladder, where they can cause infection [Reference Yamamoto6, Reference Gruneberg7]. The most common risk factors for UTI in young women include sexual intercourse and spermicide use [Reference Hooton8]. The mechanics of sexual intercourse aid in moving ExPEC from the intestine to the urethra, where the bacteria can ascend to the bladder, kidneys or move to the bloodstream [Reference Brown and Foxman9–Reference Kelsey11]. ExPEC infections are thought to be sporadic in nature and caused by a diverse collection of E. coli, which tend to possess specific virulence genes [Reference Shrikhande, Chande and Pathak12].
Over the past few decades, clusters of community-acquired, extraintestinal infections arising from genetically related groups (or clonal groups) have been documented in several countries including England [Reference Phillips13], Canada [Reference Pitout14], Denmark [Reference Olesen15] and the USA [Reference Manges16]. E. coli associated with these clusters or outbreaks were often recognized due to an unusual antimicrobial resistance phenotype or serogroup, not typically associated with such infections. The source and transmission routes for these strains have not been identified, but speculation has grown that these outbreaks may occur as the result of food or other environmental contamination [Reference Phillips13–Reference Manges16]. Moreover, these outbreak-associated ExPEC strains tend to exhibit multidrug resistance and many have been associated with severe infections [Reference Johnson17–Reference Manges19].
Increasing antimicrobial resistance in ExPEC isolates has been hypothesized to be due to the increased use of antimicrobials in human medicine. However, recent studies suggest that the development of and selection for antimicrobial-resistant E. coli may also result from the use of antimicrobial agents in food animal production for therapy, prevention and control of diseases, and to promote growth [Reference Aarestrup20]. There is evidence to suggest that the increased selective pressure on the commensal microbiota of these animals has induced resistance that may be transferred to humans, potentially via consumption of contaminated meat [Reference Aarestrup20]. Various studies have identified an abundance of antimicrobial-resistant strains of E. coli in retail meats, specifically poultry [Reference Johnson21–Reference Vincent24]. Some food isolates have been found to be indistinguishable from human clinical isolates [Reference Johnson22, Reference Vincent24]. The existence of ExPEC-associated outbreaks and the new evidence linking ExPEC in food reservoirs to human infections suggests that the epidemiology of ExPEC infections may be characterized by two models: the endemic model, where infections are caused by a range of diverse, primarily human-adapted E. coli and the epidemic model, where specific strains of E. coli found in food animal reservoirs are transmitted via food to humans periodically, as in the case of the observed outbreaks. An examination of the scientific literature for epidemiological evidence for these two models motivated this systematic review.
In this review, two types of studies were identified: (i) reports of potential outbreaks of ExPEC infections over the past 30 years; and (ii) studies conducted from the 1950s to the present, which were primarily designed to characterize ExPEC isolates (‘non-outbreak studies’) causing infections including community-acquired UTIs and other extraintestinal infections. First, epidemiological details were summarized from all identified outbreak reports, including documentation of any evidence for a food or environmental source. The seasonality of the outbreaks was also examined. Second, E. coli serogroup was identified as a marker of temporal changes in the epidemiology and distribution of ExPEC over time since its application is standard and has been used consistently in many studies since the late 1940s.
METHODS
Outbreak studies
A systematic literature review was designed to retrieve published articles of ExPEC outbreak reports from Medline from 1950 to July 2009. Published articles concerning confirmed or suspected outbreaks, as defined by the authors, of extraintestinal infections emerging from the community in otherwise healthy individuals were reviewed. All searches were performed with the assistance of an experienced librarian. The data extracted from the articles included year of outbreak, location, whether the illness associated with the infection was acquired in the community, hospital or a combination of both, the diseases associated with the outbreak, the observation period, period of peak numbers of cases, number of people and isolates used in the study, age range, the sex of those studied and the serogroup, serotype and/or multilocus sequence type (MLST) of the epidemic strain. Where possible, a designation for an E. coli strain is provided, which includes the serogroup or serotype and MLST, for example E. coli serotype O25:H4-ST131. Structured interviews were conducted with the authors of some studies to complete missing information and to identify additional studies.
Exclusion criteria for outbreak studies
Studies were excluded if they (i) reported outbreaks occurring exclusively in a healthcare setting; (ii) reported outbreaks associated exclusively with gastroenteritis or involving pathogens other than E. coli; (iii) reported outbreaks associated with animals; (iv) involved only a few case-reports; and (v) reported in languages other than French and English.
Non-outbreak studies
A similar review of ExPEC non-outbreak studies was designed using ‘human’, and English and French language limits. These studies contained information on E. coli typically responsible for extraintestinal infections occurring in the community in otherwise healthy individuals. The data extracted from the articles included study observation period, location, whether the infection was acquired in the community, hospital or both, infection type, the number of people and isolates included, age range, the sex of those studied, the number of O-antisera used and the identified E. coli serogroups. Studies that did not provide the number of antisera used for testing were included if a reference laboratory was cited, or if the number of non-typable isolates was low. This criterion was used to ensure that the isolates were completely characterized according to serogroup. The proportion of each serogroup was calculated using the total number of isolates in the study as the denominator, unless otherwise indicated. A weighted average for each serogroup was estimated using the inverse of the number of isolates in the study. In some cases, values were collapsed (e.g. if UTI and pyelonephritis serogroups were reported separately), and thus may differ from those reported in the articles.
Exclusion criteria for non-outbreak studies
Published articles were excluded if they (i) focused exclusively on complicated infections (i.e. pregnant women, children or infants, the institutionalized elderly, those with underlying conditions such as diabetes or, hospitalized patients, catheterized patients, patients with urological abnormalities and chronic UTIs [Reference Foxman5]), (ii) reported serotyping results based on fewer than 50 O-antisera, (iii) included results from intervention studies or randomized control trials, (iv) focused exclusively on bacteriuria, or (v) were published in a language other than English or French. Only a single article was included if multiple articles based on the same study population were identified. In both searches, studies that did not clearly conform to our criteria were discussed and evaluated by both reviewers.
RESULTS
ExPEC outbreaks
Twelve ExPEC outbreaks studies were included in this review (see Fig. 1). ExPEC outbreaks were identified in UK, Denmark, USA, Spain, Canada, Croatia, and Portugal. The earliest outbreak detected occurred in 1986 and the latest in 2008. The illnesses associated with these outbreaks were primarily related to the urinary tract but were also associated with blood(+stream) and other infections. Both males and females, across a wide age range, were affected. The incidence of the outbreak strains in these reports varied from <1% to 26%, after excluding studies with exclusively antimicrobial-resistant isolate samples. The most common serogroups associated with ExPEC outbreaks included O15 and O25. The articles are summarized in Table 1 and below.
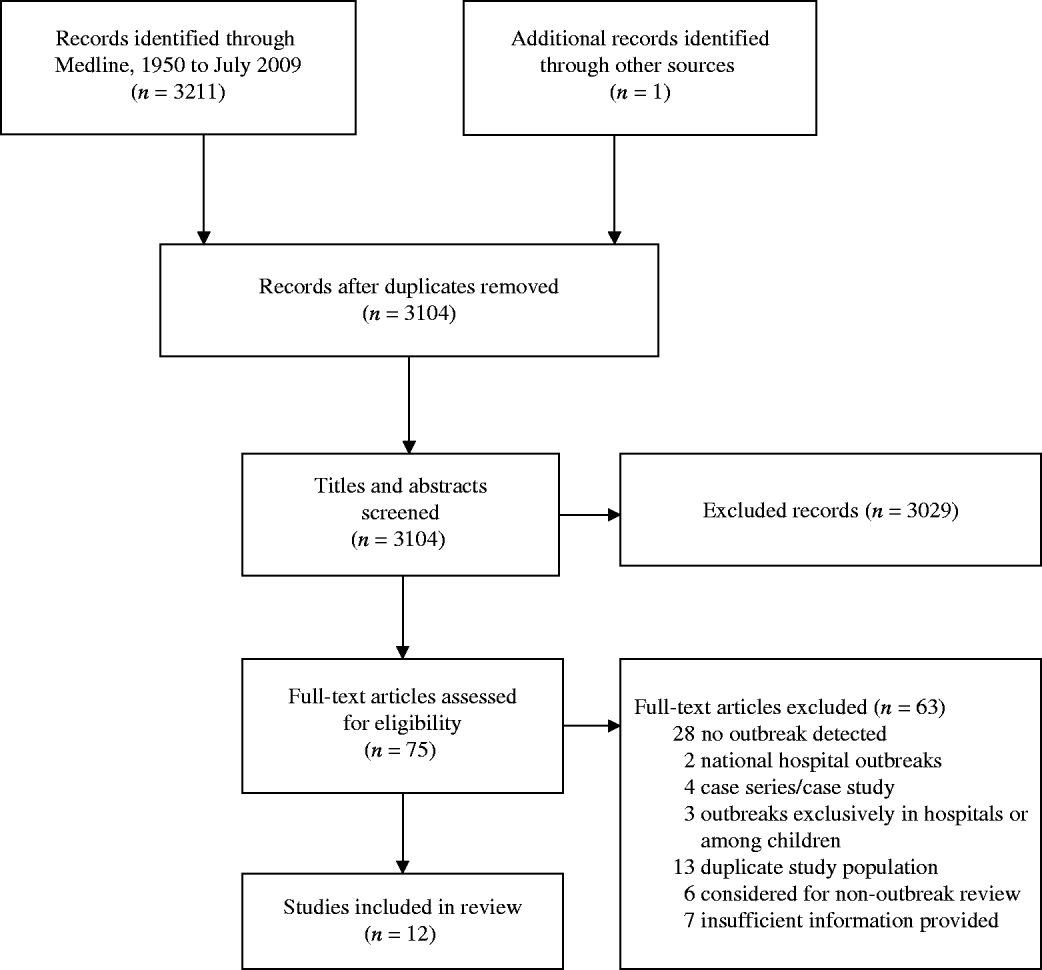
Fig. 1. The selection process of outbreak ExPEC articles into the review. Flow chart adapted from Moher et al. [Reference Moher77].
Table 1. Reported outbreaks of community-acquired Escherichia coli causing human extraintestinal infections
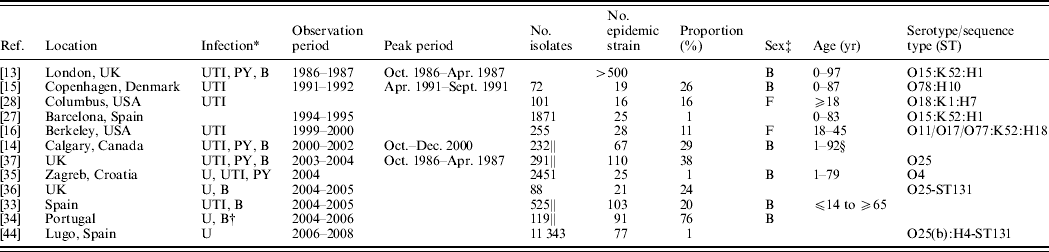
* B, Isolates recovered from blood samples, bacteraemia cases or sepsis cases; U, Isolates recovered from urine samples; UTI, isolates recovered from cases of cystitis or UTIs; PY, isolates recovered from pyelonephritis cases.
† E. coli was also recovered from wounds, ascitic fluid, sputum, gastric fluids, bronchioaveolar lavage and secretions.
‡ M, male; F, female; B, both male and female.
§ Dr J. Pitout, personal communication.
|| Indicates that the proportion is estimated based on a sub-sample of ESBL-producing E. coli and therefore does not reflect the overall proportion.
West Lambeth, London, 1986–1987, serogroup O15:K52:H1
An uncommon E. coli strain was responsible for a community outbreak between October 1986 and October 1987 in Southeast London, England, involving urinary, bloodstream and other infections [Reference Phillips13]. The strain was first recognized due to the unusual antimicrobial pattern which included resistance to ampicillin, chloramphenicol, streptomycin, sulphonamide, tetracycline and trimethoprim. During the outbreak, this strain was associated with 15% of all urinary isolates observed over the year in the region. Moreover, 84/819 patients grew the epidemic E. coli, identified by serotyping, from stool samples. This E. coli serotype caused 29 cases of septicaemia during the outbreak, while it had caused only 16/674 septicaemia cases over the previous 17 years [Reference Phillips13]. Following this reported outbreak in West Lambeth, similar findings were documented in Roehampton [Reference Wright and Perinpanayagam25] and South London [Reference Waghorn, Kelly and Gibbins26]. In addition, a study in Barcelona, Spain found the epidemic strain in 1·3% of more than 1000 urine and blood samples received between June 1994 and May 1995, nearly a decade after the London outbreak [Reference Prats27].
Ohio, serogroup O18:K1:H7
In a study of the nutritional and osmotic requirements of clinical E. coli isolates, a group of strains requiring nicotinamide for growth and exhibiting the O18:K1:H7 serotype was isolated from 16/101 consecutive urine samples from young women with cystitis and 5/100 stool samples from healthy subjects in Ohio [Reference Kunin28]. The O18:K1:H7 serotype was found to be similar to that associated with neonatal meningitis and sepsis. Indistinguishable RAPD patterns were also observed between neonatal meningitis, urine and faecal isolates belonging to O18:K1:H7 [Reference Johnson, Delavari and O'Bryan29]. This serotype of E. coli is suspected of colonizing the vagina or intestines of women and may consequently enter the bladder, causing cystitis, or may be transferred to infants during or immediately following birth [Reference Kunin28].
Copenhagen, 1991, serogroup O78:H10
In 1991, a year-long outbreak of multidrug-resistant E. coli O78:H10 occurred in the Greater Copenhagen region of Denmark [Reference Olesen15]. Infections with the epidemic serotype occurred in 15 females and three males. All isolates were recovered from urine specimens and 13 cases were related to UTI. According to the authors, at least 14 cases were community-acquired infections [Reference Olesen15]. The epidemic strain was resistant to ampicillin, chloramphenicol, tetracycline, streptomycin, sulphonamides and trimethoprim. A single O78:H10 outbreak isolate was identified from among 500 stool samples collected over an 8-month period. E. coli O78 was known to cause septicaemia in calves, but had not previously been reported to cause human disease. Only 30 O78:H10 isolates were identified during a retrospective study of isolates collected from around the world between 1956 and 1990 by the WHO International Escherichia and Klebsiella Centre at the Statens Serum Institut. Although, epidemiological information was not available for this outbreak, the authors concluded that the outbreak was most likely associated with foodborne transmission; however, a vehicle was not identified.
Berkeley, California, 1999–2001, serogroup O11/O17/O73/O77:K52:H18-ST69
During a study analysing urine samples from women in three different communities in the USA, a multidrug-resistant E. coli clonal group, designated clonal group A (CgA), caused one-third to one-half of all TMP-SMZ-resistant [Reference Manges16] cases of UTI. Forty-one percent of the California CgA isolates were indistinguishable by XbaI PFGE fingerprinting. CgA members belonged to serotypes O11 and O77:K52:H18 at the California study site; and were later found to belong to serogroups O17 and O73 [Reference Johnson17] and exhibit MLST ST69 [Reference Tartof30]. In addition to TMP-SMZ, this clonal group also exhibited resistance to ampicillin, tetracycline, chloramphenicol, and streptomycin. A peak period could not be ascertained due to the 4 months' duration of the study. Dissemination throughout the community was observed through the recovery of CgA from stool samples from 15/41 healthy subjects during the same study period [Reference Manges16].
A second cross-sectional study was conducted 1 year later to evaluate the hypothesis that CgA was associated with an outbreak of UTI [Reference Manges31]. In the second study, only four (11%) TMP-SMZ-resistant CgA isolates were identified vs. 23 (49%) in the first study (P<0·001). The temporal decline in UTI cases associated with CgA provided further evidence that CgA may have caused a community outbreak of UTI. Alternatively, the decline may be indicative of a decrease in the endemicity of CgA in UTIs (J. R. Johnson, personal communication). Six other clonal groups were responsible for 32% of TMP-SMZ-resistant UTIs during the observation period [Reference Manges31]. As with CgA, members of these other clonal groups exhibited closely related PFGE patterns and were recovered from unrelated women [Reference Manges31]. The fluctuation of other E. coli clonal groups in this community suggested that a large proportion of community-acquired UTIs may be caused by E. coli disseminated from one or more point sources. However, epidemiological evidence indicating a source was not available.
Outbreaks associated with extended spectrum β-lactamase (ESBL)-producing E. coli
The production of β-lactamases by ExPEC has complicated the treatment of UTIs and other infections. These newly emerging pathogens have developed resistance to commonly used antibiotics, such as cephalosporins. Such organisms have been found in healthcare facilities since the 1980s, but are now of even greater concern due to the increase in incidence in the community, especially the CTX-M subtypes [Reference Pitout32]. Dissemination of related ESBL-producing strains have also been documented in Canada [Reference Pitout14], Spain [Reference Oteo33], Portugal [Reference Mendonca34], Croatia [Reference Vranes35] and the UK [Reference Lau36, Reference Woodford37]. ESBL-producing E. coli O25:H4-ST131 is one clone which appears to have spread widely and has been responsible for several outbreaks.
Calgary, Alberta, Canada, 2000–2001
Between April 2000 and December 2001, Pitout et al. reported a community-wide outbreak in Calgary, Alberta of a CTX-M-14 β-lactamase-producing E. coli clonal group, with a peak in cases falling approximately between October and December 2000 [Reference Pitout14]. This clonal group, designated CTX-M-14A and defined by closely related XbaI PFGE patterns, caused infections in 59 patients, predominately of community onset (80%), all identified from urine specimens. The epidemic strain was associated with phylogenetic group D [Reference Pitout38]. This was the first reported outbreak associated with community-acquired ESBL-associated infections. The authors of this report also suggest the possibility of a common food or environmental source; however, epidemiological information to test this hypothesis was unavailable. An increase in CTX-M-14 isolates was also observed in the same region in 2003 [Reference Pitout39] and had also spread to Edmonton, Canada (J. D. Pitout, personal communication).
Madrid, Spain, 2004–2005
During a surveillance study of ESBL-producing E. coli strains which were resistant to cefotaxime and ceftazidime, a multidrug resistant, epidemic strain was identified in 103/525 isolates collected from mostly urine, blood and wound samples from January 2004 to August 2005 [Reference Oteo33]. The outbreak strain, identified by XbaI PFGE, most commonly exhibited resistance to ciprofloxacin, gentamicin, tobramycin, ampicillin, amoxicillin-clavulanic acid and TMP–SMX.
Portugal, 2004–2006, serogroup O25:H4-ST131
Of 181 ESBL-producing E. coli isolates collected and analysed from mainly urine specimens between March 2004 and March 2006 in Portugal, 91 comprised a group of related or indistinguishable isolates by XbaI PFGE [Reference Mendonca34]. The majority of this cluster produced CTX-M-15 enzymes and 58% of these isolates were recovered from outpatients. The predominant resistance profile included resistance to ampicillin (±sulbactam), aztreonam, piperacillin, ticarcillin, ciprofloxacin, gentamicin and tobramycin. A small sample of these isolates were evaluated for serogroup, MLST (ST131) and phylogroup (B2) [Reference Nicolas-Chanoine40].
Zagreb, Croatia, 2004, serogroup O4
A 5-month study in Croatia, between January and May 2004, revealed a clonally related cluster defined by XbaI PFGE of 25 ESBL-producing E. coli isolates recovered from non-hospitalized patients [Reference Vranes35]. These isolates belonged to serogroup O4 and exhibited a common pattern of resistance to gentamicin, amikacin, netilmicin and TMP-SMX.
United Kingdom, 2003–2004, serogroup O25-ST131
Two clonal groups A and D, responsible for a community and hospital outbreak, were identified during 2003–2004 in the UK from urine and blood samples from more than 200 cases [Reference Woodford37, Reference Karisik41, Reference Warren42]. Both strains were shown to be E. coli O25, most produced CTX-M enzymes [Reference Woodford37] and were resistant to quinolones and trimethoprim [Reference Warren42]. Strains A and D belonged to phylogenetic B2 [Reference Karisik43], were closely related by PFGE analysis and belonged to ST131. Faecal carriage of strains was found in community and hospital diarrhoeal samples [Reference Warren42]. Certain outpatients were interviewed but no association with food or retail outlets was noted [Reference Warren42].
A similar outbreak was identified in northwest England during October 2005 [Reference Lau36]. These isolates were recovered from blood and urine samples were also found to belong to ST131.
Lugo, Spain, 2006–2008 O25:H4-ST131
In a study involving ESBL-producing isolates from Lugo, Spain, a cluster of strains belonging to serogroup O25b:H4 and ST131 was identified between February 2006 and May 2008 [Reference Blanco44]. All isolates in this group belonged to phylogenetic group B2 and demonstrated resistance to ciprofloxacin, TMP-SMX and tobramycin. The epidemic strain was isolated mainly from urine samples. CTX-M-15-producing isolates accounted for about 20% of all ESBL-producing isolates during 2006–2008.
ExPEC non-outbreak studies
After extracting information from full-article texts, 28 eligible non-outbreak studies were included in this review (see Fig. 2). A summary of the studies and serogroups identified in the ExPEC non-outbreak studies is presented in Table 2. These studies were conducted in USA, UK, Spain, Ireland, Denmark, The Netherlands, Australia, New Zealand, South Africa, China, Japan, and India between 1960 and 2001. The studies are presented from earliest sampling period to most recent period; if this information was not reported, publication date was used. Both men and women were included in the studies and a wide range of extraintestinal infections including cystitis, pyelonephritis, bacteraemia and prostatitis were observed. Eleven studies used samples from outpatients only, while eight used a combination of both in- and outpatient samples. Nine studies did not specify the population type. It is possible that some of the non-outbreak studies, particularly the earlier studies, contained unrecognized outbreaks. The serogroups most commonly associated with UTIs were O1, O2, O4, O6, O7, O8, O16, O18, O25, and O75 [Reference Gruneberg7, Reference Orskov45]. The weighted proportion of these serogroups varied from 1% to 15%. Serogroup O6 occurred most frequently, ranging in incidence from 4% to 30%, and was identified in every study. Serogroups O2, O4, and O75 were the next most common serogroups reported and were identified in almost all studies (Table 2). Our results are consistent with the observed distribution of ExPEC recovered from infections from other studies [Reference Johnson46, Reference Vosti47], although proportions may vary temporally and geographically [Reference Gruneberg7, Reference Mabeck, Orskov and Orskov48].
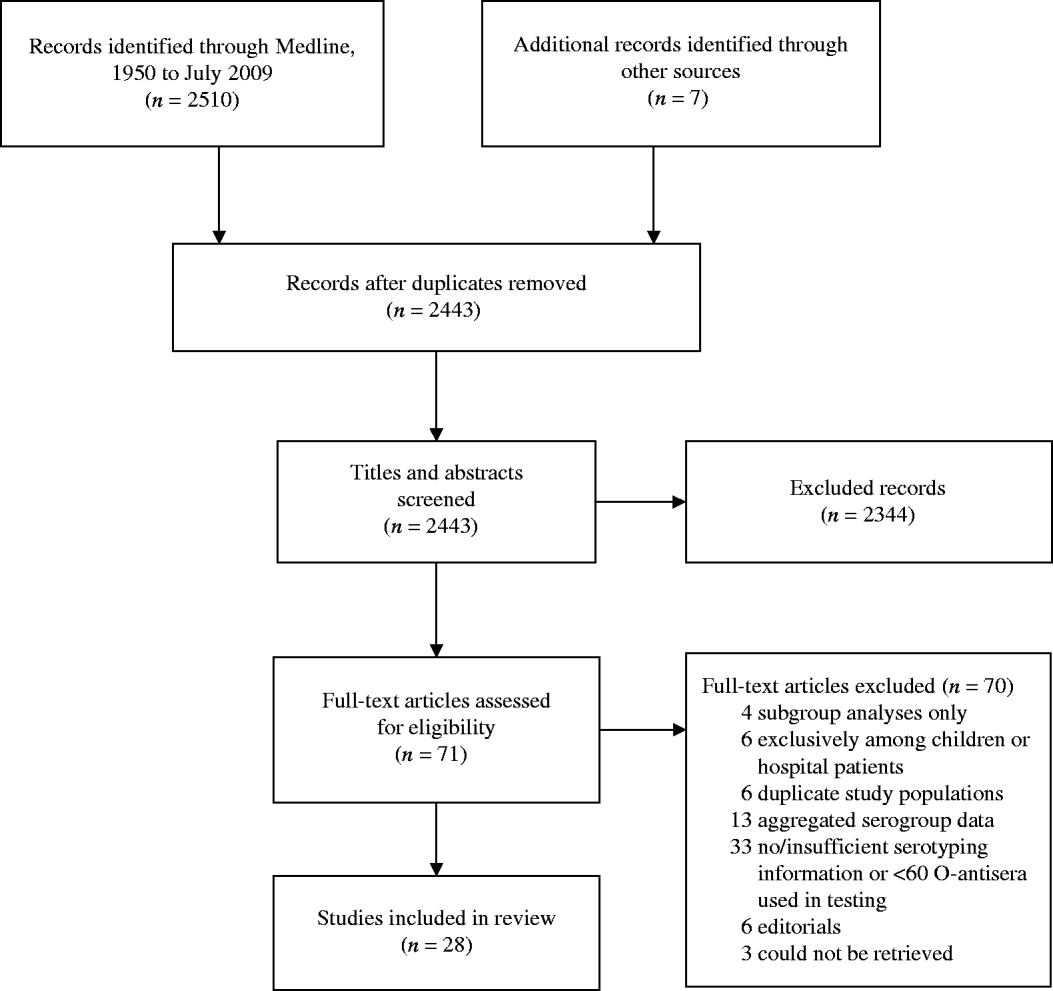
Fig. 2. The selection process of non-outbreak ExPEC articles into the review. Flow chart adapted from Moher et al. [Reference Moher77].
Table 2. Reported serogroups of Escherichia coli causing human extraintestinal infections: non-outbreak studies
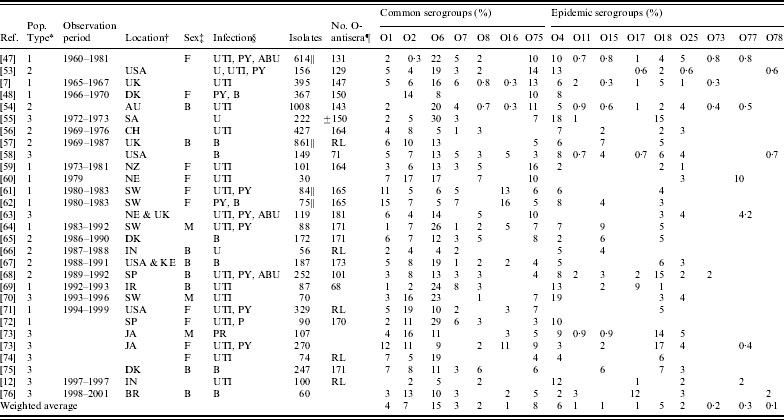
* Population type: 1, community-acquired infections; 2, community- and hospital-acquired infections; 3, patient population type not reported.
† AU, Australia; CH, China; UK, England; DK, Denmark; SP, Spain; NE, The Netherlands; SW, Sweden; JA, Japan; FN, Finland; CR, Croatia; CA, Canada; PR, Portugal; IN, India; BR, Brazil; KE, Kenya; IR, Iran; SA, South Africa; NZ, New Zealand.
‡ M, male; F, female; B, both male and female.
§ B, Isolates recovered from blood samples, bacteraemia cases or sepsis cases; U, isolates recovered from urine samples; UTI, isolates recovered from cases of cystitis or UTIs; PY, isolates recovered from pyelonephritis cases; PR, isolates recovered from prostatitis cases; ABU, isolates recovered from asymptomatic bacteriuria cases.
|| The denominator used for calculations may differ from the number of isolates tested. For Vosti [Reference Vosti47], the denominator is 614 due to missing information from 291 patients; for Grandsen et al. [Reference Gransden57], the denominator is 861 which is the number of patients studied; for Sandberg et al. [Reference Sandberg61], the denominator is 84 (only non-pregnant PY and UTI patients included); for Otto et al. [Reference Otto62] the denominator is 75 (92 minus complicated cases, including diabetic patients).
¶ RL was used when serotyping was done at a reference laboratory and was assumed to use the entire set of O-antisera present at the time of the study.
DISCUSSION
Extraintestinal infections caused by E. coli are believed to be sporadic, opportunistic infections. This notion, however, is being challenged by the recognition of clusters of infections caused by indistinguishable or closely related E. coli, occurring in specific geographic locations over short periods of time. This is the first epidemiologial review and comparison of published reports describing ExPEC-associated outbreaks and reports describing community-acquired infections caused by endemic or sporadic ExPEC. The existence of ExPEC outbreaks appears well supported. It is likely that the occurrence of these outbreaks has been substantially under-reported; however, the availability of inexpensive genotyping assays has probably led to an increase in the number of ExPEC outbreak reports since the 1990s.
Apart from serogroups O4, O18 and O25, the ExPEC outbreak-related serogroups were relatively uncommon in reports of sporadic ExPEC-associated infections. Moreover, the proportion of infection with an outbreak-associated serogroup was notably higher during the outbreak periods (1–26%, excluding reports with exclusively antimicrobial-resistant isolate samples) than during non-outbreak periods (weighted proportions <1–15%). Furthermore, serogroup O25, which was commonly associated with endemic ExPEC infections (weighted proportion 2%), has recently been associated with the emergence of ESBL-producing E. coli, and was responsible for several outbreaks [Reference Pitout14, Reference Oteo33–Reference Vranes35, Reference Woodford37, Reference Warren42, Reference Blanco44]. Although some of the epidemic serogroups or serotypes were detected in the non-outbreak studies, the proportion of these serogroups during an epidemic was higher. Most outbreaks involved community-acquired UTIs, and females were disproportionately affected. The age range of infected persons was broad. The duration of the outbreaks was difficult to assess since many studies inadvertently discovered the outbreak or were retrospective in design.
ExPEC infections may follow a seasonal pattern, similar to sexually transmitted infections [Reference Freeman, Anderson and Sexton49]. The frequency of UTIs and bloodstream infections have been shown to peak during the warmer seasons [Reference Al-Hasan50, Reference Anderson51]. It has been suggested that warmer temperatures may enhance growth of these types of E. coli in food or environmental sources, and may promote the expression of certain virulence factors [Reference Freeman, Anderson and Sexton49]. Only four of the outbreak studies reported information on the timing of the peak period of infections; three of these four reports indicated an observed peak in the winter months. Due to limited data on reported peak periods, conclusions vis-à-vis seasonality could not be drawn.
The source of the E. coli involved in these outbreaks remains unclear, although several reports reviewed suggest a food or shared community exposure. Although no source was identified in any of the outbreaks, there has been evidence to support food as a potential reservoir for ExPEC. Isolates cultured from retail meats and other food items have been found to resemble human clinical isolates from urine and faeces [Reference Johnson22, Reference Vincent24]. Further, during an outbreak involving gastroenteritis due to Salmonella, a bacterium known to be contracted via undercooked meat and other food items, at a camp in Girona, Spain, investigators detected genetically related extended-spectrum cephalosporin-resistant E. coli. Clones A and B were confined to those individuals with Salmonella infections. Those infected had no previous contact before camp yet lived together and shared a water and food supply during camp [Reference Prats52]. Human contamination might also have contributed to the spread of the strains, yet food handlers surveyed during the camp outbreak did not possess the epidemic strains of E. coli, nor was any transmission detected in the households of those colonized [Reference Prats52]. Several of the outbreak studies also included a stool survey which allowed for the detection of the outbreak strains circulating in healthy community members [Reference Manges16, Reference Kunin28, Reference Prats52]. Together, the evidence points to food as a potential reservoir of ExPEC, which would lead to intestinal colonization with these strains.
There are several limitations to this review. Incomplete data collection and reporting in the outbreak studies, particularly related to epidemiological information including complete dates, subject sample size, and precise sex and age distributions, was problematic. The outbreak studies reported were often conducted once the outbreak was underway and may not reflect the entire time the epidemic strain was circulating in the community; moreover, many reports did not continue surveillance or conduct repeated sampling over time. The outbreaks were identified primarily due to an unusual serogroup, antibiotic profile or growth conditions exhibited by the outbreak strain, suggesting that the characteristics of the outbreak strains are biased and outbreaks due, for example, to antibiotic-susceptible ExPEC, would be under-reported. The association between infection with an outbreak strain and the development of more severe disease may be biased as laboratory or diagnostic testing occurs more frequently with severe infections, treatment failures or recurrent infections. Serogroup was used a marker of E. coli dynamics over time in studies of extraintestinal infections. Serogroup has been consistently collected in many studies over time and therefore allows for temporal analysis of studies of extraintestinal infections from 1950 onwards. However, specific serogroups will encompass multiple genotypes; therefore serogroup is an imperfect biomarker for our comparisons. Only studies with O-antisera testing panels containing >50 serogroups were included to ensure representation of most circulating serogroups. Some studies may have included recurrent infections or collected multiple samples for a single person; this may have led to over-estimation of certain serogroups in the non-outbreak studies.
Many questions remain unanswered about the epidemiology of ExPEC and their associated infections. The origin and mode of transmission of these types of E. coli has not yet been confirmed but evidence suggests foodborne dissemination [Reference Manges16, Reference Vincent24, Reference Prats52]. Despite limitations in reporting, this review highlights the existence of outbreaks associated with specific types of ExPEC and illustrates the differences in serogroup in ExPEC associated with outbreaks and ExPEC that have been responsible for sporadic disease over the past several decades. The frequency and magnitude of ExPEC outbreaks remains unclear, but their existence is strongly supported.
ACKNOWLEDGEMENTS
We thank Gurjit Toor for her assistance in gathering and summarizing some articles for this study and the members of the Manges Laboratory. We are also grateful to the authors of key research reports who agreed to complete the structured interviews.
DECLARATION OF INTEREST
None.






