Background
The human microbiome is the ecosystem of microorganisms that inhabit the human body. The majority of these microbes are found in the human intestines, where they play important roles in epithelial barrier function, fermentation of dietary fibre, synthesis of vitamins, calibration of the immune system, and defence against pathogens.Reference Fujimura, Slusher, Cabana and Lynch1 This community has also been found to play some surprising roles in angiogenesis,Reference Schirbel, Kessler and Rieder2,Reference Stappenbeck, Hooper and Gordon3 brain development,Reference Ceppa, Mancini and Tuohy4–Reference Diaz Heijtz, Wang and Anuar6 and behaviour.Reference Diaz Heijtz, Wang and Anuar6,Reference Clarke, O’Mahony, Dinan and Cryan7
Given the various functions of the human-associated microbiota, it is unsurprising that aberrations in this community (dysbiosis) have been associated with a range of non-communicable diseases (NCDs), including obesity,Reference Boulange, Neves, Chilloux, Nicholson and Dumas8–Reference Moreno-Indias, Cardona, Tinahones and Queipo-Ortuno12 diabetes,Reference Moreno-Indias, Cardona, Tinahones and Queipo-Ortuno12,Reference Tilg and Moschen13 inflammatory bowel disease,Reference Papa, Docktor and Smillie14,Reference Ni, Wu, Albenberg and Tomov15 metabolic syndrome,Reference Parekh, Balart and Johnson16,Reference Chassaing and Gewirtz17 some types of cancer,Reference Dahmus, Kotler, Kastenberg and Kistler18–Reference Flemer, Lynch and Brown20 asthma,Reference Abrahamsson, Jakobsson, Andersson, Bjorksten, Engstrand and Jenmalm21–Reference Thorburn, McKenzie and Shen23 allergy,Reference Bjorksten, Sepp, Julge, Voor and Mikelsaar24–Reference Subbarao, Anand and Becker26 non-alcoholic fatty liver disease (NAFLD),Reference Kolodziejczyk, Zheng, Shibolet and Elinav27 and even certain neuropsychiatric disorders.Reference Warner28 NCDs such as these are a significant and increasing burden on our health systems and on society. The rise of NCDs, which has been described as ‘a public health emergency in slow motion’,Reference Ki-moon29 has coincided with environmental, medical, and lifestyle changes that are known to impact on the human microbiome. Such practices include antibiotic use, dietary antimicrobials, increased hygiene, formula feeding, and reduced exposure to animals and farming environments.Reference Bailar and Travers30–Reference Thorsen, McCauley and Fadrosh33
The developmental origins of health and disease (DOHaD) paradigm provides a framework through which to understand programming of disease by the human microbiome. The DOHaD theory states that exposures during the highly plastic period of early life can programme later-life disease risk. The human microbiome is seeded, established, and stabilises in early life.Reference Hornef and Haller34,Reference Stewart, Ajami and O’Brien35 During the first 2–3 years of life, the human gut microbiome is highly dynamic and is shaped by numerous environmental factors.Reference Stewart, Ajami and O’Brien35 Early life (from gestation to early childhood) appears to be a critical window for microbiome-driven programming of NCDs. Here, the evidence that certain NCDs have an early-life microbiome origin is reviewed, with a focus on insults that can disrupt early-life establishment of the human microbiome.
The early-life microbiome contributes to later-life disease
The microbiome is established in a clear, stepwise manner in the first years of life.Reference Stewart, Ajami and O’Brien35–Reference Backhed, Roswall and Peng38 At birth, the human gut microbiome is highly dynamic, with a low biomass, a low level of intra-individual (alpha) diversity, and a high level of inter-individual (beta) diversity.Reference Ferretti, Pasolli and Tett39,Reference Yassour, Jason and Hogstrom40 At around 1 year of age, the gut microbiome shifts from a developmental stage to a transitional phase.Reference Stewart, Ajami and O’Brien35 During this period, the composition of the microbiome changes, as alpha diversity increases and beta diversity decreases. Finally, 2–3 years after birth, the gut microbiome matures to a stable, adult-like phase.Reference Stewart, Ajami and O’Brien35 A range of chronic health conditions have been shown to be preceded by or associated with dysbiosis during this early-life critical window of microbiome development.
Asthma
Evidence from longitudinal cohort studies suggests that a change in the early-life intestinal microbiota precedes development of wheeze and asthma in childhood. Stokholm et al. reported an association between an immature gut microbiome at 1 year of age and development of asthma at 5 years of age in a cohort of 690 children.Reference Stokholm, Blaser and Thorsen41 Importantly, this association was only evident in the children of asthmatic mothers, suggesting that the double hit of genetic predisposition and lack of microbial stimulation in infancy triggers asthma development. In another study of 1176 children, faecal colonisation with Clostridium difficile at 1 month of age was correlated with wheezing throughout the first 6–7 years of life and asthma at 6–7 years of age.Reference van Nimwegen, Penders and Stobberingh42 Similarly, Fujimura et al. analysed stool samples from 298 infants aged 1–11 months and found that infants with a lower relative abundance of particular bacteria (such as Bifidobacterium, Akkermansia, and Faecalibacterium), and a higher relative abundance of the fungal genera Candida and Rhodotorula had an increased risk of asthma at 4 years of age.Reference Fujimura, Sitarik and Havstad43 In a smaller study of just 47 infants, children who developed asthma by 7 years of age (n = 8) showed decreased gut microbiome diversity at 1 week and 1 month of age, but not at 12 months.Reference Abrahamsson, Jakobsson, Andersson, Bjorksten, Engstrand and Jenmalm21 However, with such a low number of participants, and such a high level of inter-individual variation at these time points, these data should be interpreted with caution. Antibiotic use in the first 2 years of life increases the risk of wheezeReference Kummeling, Stelma and Dagnelie44 and asthmaReference Hoskin-Parr, Teyhan, Blocker and Henderson45 in later childhood in a dose-dependent manner. Together these studies demonstrate that dysbiosis of the intestinal microbiome in the first year of life underpins later-life risk of asthma.
Further evidence from animal studies supports the notion that the early-life microbiome plays a role in asthma development. In germ-free mice, invariant natural killer T cells accumulate in the colonic lamina propria and lung, leading to increased disease severity in an ovalbumin-challenged mouse model of allergic asthma.Reference Olszak, An and Zeissig46 Colonisation of these mice with conventional murine microbiota protects them from asthma, but only if performed in the neonatal period. Arrieta et al. found four bacterial genera (Faecalibacterium, Lachnospira, Rothia, and Veillonella) were decreased in 3-month-old stool samples from CHILD study participants who went on to develop wheeze and asthma in later life.Reference Arrieta, Stiemsma and Dimitriu22 Importantly, when these four taxa were inoculated into pregnant germ-free mice, their adult progeny were protected from airway inflammation.
There is evidence to suggest that the mechanism by which the early-life gut bacteria protect children from developing asthma is via their production of short-chain fatty acids (SCFAs). SCFAs are immune-modulating bacterial metabolites which are produced in the intestines as a result of fibre fermentation and are distributed systemically. Thorburn et al. found that, in a mouse model of allergic airway inflammation, maternal gestational high-fibre diet or supplementation with the SCFA acetate altered the maternal microbiome and subsequently protected the offspring from asthma.Reference Thorburn, McKenzie and Shen23 The offspring of the high-fibre diet/acetate-supplemented dams showed an increase in T-regulatory cell number and function, as well as epigenetic changes in the Foxp3 promotor. Further, high serum acetate levels in pregnant women correlates with reduced risk of wheeze in their offspring in the first year of life.Reference Thorburn, McKenzie and Shen23 Similarly, high intestinal acetate at 3 months of age,Reference Arrieta, Stiemsma and Dimitriu22 and propionate and butyrate at 12 months of ageReference Roduit, Frei and Ferstl47 in infants correlates with a decreased risk for wheeze and asthma in later life. These studies demonstrate the importance of the early-life gut bacteria in shaping appropriate immune functions via the actions of SCFAs.
Allergy and atopy
Dysbiosis of the intestinal microbiome has been shown to precede development of allergy and atopy. In mice, neonatal antibiotic exposure alters the intestinal microbiota and enhances food allergen sensitisation.Reference Stefka, Feehley and Tripathi48 In the CHILD study cohort, an increased Enterobacteriaceae/Bacteroidaceae ratio at 3 months and 1 year of age was associated with increased food allergen sensitisation at 1 year.Reference Azad, Konya and Guttman49 In the KOALA study cohort, Escherichia coli colonisation in stool samples taken at 1 month of age was associated with eczema diagnosis by 2 years of age in a dose-dependent fashion, while C. difficile colonisation was associated with both eczema and allergic sensitisation.Reference van Nimwegen, Penders and Stobberingh42,Reference Penders, Thijs and van den Brandt50 In this same cohort, the effect of intestinal colonisation with E. coli on eczema development was modified by genetic variations in CD14 (cluster of differentiation 14) and TLR4 (Toll-like receptor 4), suggesting that host gene–microbiota interactions determine atopy development in early life.Reference Penders, Thijs and Mommers51 In a study of 138 infants, discordant temporal development of Enterobacteriaceae and Parabacteroides spp. and decreased abundance of Eubacterium spp. and Anaerostipes spp. over the first 26 weeks of life were observed in those that were diagnosed with eczema by 18 months of age (n = 52).Reference Wopereis, Sim, Shaw, Warner, Knol and Kroll52 These infants also had decreased levels of the SCFA butyrate in their stool in the first 26 weeks of life. This finding has been expanded upon in another study showing that gut dysbiosis and decreased stool SCFA levels at 3 months of age correlated with atopy at 1 year of age.Reference Stiemsma, Arrieta and Dimitriu53 Children with the highest levels of stool butyrate have been shown to have the lowest risk of food allergy and allergic rhinitis in later life.Reference Roduit, Frei and Ferstl47 These data suggest that early-life dysbiosis drives alterations in the SCFA profiles of infants, which may underpin the risk of atopy and allergy.
Obesity
There is a wealth of literature implicating the human gastrointestinal microbiome in the aetiology of obesity. An obesogenic (‘Western’) diet causes the microbiome to shift to an obesogenic state,Reference Turnbaugh, Ridaura, Faith, Rey, Knight and Gordon54–Reference Turnbaugh, Backhed, Fulton and Gordon56 while an obesogenic microbiome contributes to the development of obesity and metabolic syndrome.Reference Tseng and Wu57–Reference Cornejo-Pareja, Munoz-Garach, Clemente-Postigo and Tinahones60 Mechanistically, an obesogenic microbiome is more efficient at extracting energy from food, increasing intestinal permeability, modulating satiety hormones, increasing lipogenesis, and inducing chronic inflammation.Reference Tseng and Wu57–Reference Cornejo-Pareja, Munoz-Garach, Clemente-Postigo and Tinahones60
The rise in childhood obesity rates over the past three decades has paralleled the rise in maternal obesity. Recent models predict that more than half (57%) of today’s children will be obese by age 35.Reference Afshin and Forouzanfar61 Disruptions in the development of the early-life intestinal microbiota are associated with later-life development of obesity. Dogra et al. reported that changes in the temporal development of the gut microbiome in the first 6 months of life were predictive of adiposity at 18 months of age.Reference Dogra, Sakwinska and Soh62 Specifically, infants with high levels of Streptococcus and low levels of Bifidobacteria and Collinsella at 6 months of age showed increased adiposity at 18 months. Another prospective study of 165 children found that the composition of the gut microbiome at 10 days and 2 years was significantly associated with childhood BMI (at age 12).Reference Stanislawski, Dabelea and Wagner63 In this cohort, the gut microbiota composition at 2 years of age explained over 50% of the variation in childhood BMI. Antibiotic administration in the first year of life has been repeatedly shown to increase the risk of adiposity and obesity in later childhood.Reference Trasande, Blustein, Liu, Corwin, Cox and Blaser64–Reference Bailey, Forrest, Zhang, Richards, Livshits and DeRusso66 These observations are supported by studies in mice, which show that even sub-therapeutic doses of antibiotics in the neonatal period result in increased adiposity and metabolic hormones, as well as an increased susceptibility to a high-fat diet (HFD) in later life.Reference Cho, Yamanishi and Cox67,Reference Cox, Yamanishi and Sohn68
The maternal microbiome also appears to contribute to the risk of obesity in her offspring. Maternal antibiotic administration during the second and third trimesters increases the risk of obesity in her children at 7 years of age by 84%.Reference Mueller, Whyatt and Hoepner69 Similarly, maternal HFD during pregnancy is associated with changes in the neonatal gut microbiome independently of maternal BMI.Reference Chu, Antony and Ma70 Evidence from animal models supports the notion that dysbiosis of the maternal microbiome results in neonatal dysbiosis and obesity development. In mice, maternal HFD alters the gut microbiome in offspring, leading to the development of obesity and metabolic syndrome.Reference Paul, Bomhof, Vogel and Reimer71,Reference Wankhade, Zhong and Kang72 In a non-human primate model, maternal HFD caused changes in the composition of the neonatal gut microbiome which persist even after weaning and transfer to a control diet.Reference Ma, Prince and Bader73
Neuropsychiatric
In adults, the intestinal microbiota interact with the brain and the nervous system via their production of inflammatory and endocrine molecules and microbial metabolites.Reference Bravo, Forsythe and Chew74–Reference Dinan and Cryan78 Additionally, they are able to signal through the gut–brain axis via the vagus nerve.Reference Bravo, Forsythe and Chew74–Reference Dinan and Cryan78 Several neuropsychiatric disorders have been associated with gut dysbiosis in adults, including autism,Reference Liu, Li, Wu, Zheng, Peng and Zhou79 Parkinson’s,Reference Sampson, Debelius and Thron80 Alzheimer’s,Reference Vogt, Kerby and Dill-McFarland81 depression,Reference Valles-Colomer, Falony and Darzi82 and schizophrenia.Reference Zheng, Zeng and Liu83 Emerging evidence suggests that neuropsychiatric disorders might be programmed through early-life microbial exposure. In a cohort study of 7794 people, periconceptional genital/reproductive tract infections significantly increased the risk of schizophrenia in the offspring.Reference Babulas, Factor-Litvak, Goetz, Schaefer and Brown84 In a larger study of almost 2 million people, maternal infections during pregnancy and childhood infections during the first 13 years of life have been associated with the development of psychosis.Reference Blomstrom, Karlsson, Gardner, Jorgensen, Magnusson and Dalman85 However, in these cases, it is difficult to ascertain whether the involved mechanism is the maternal infection per se, or the inflammatory response to the infection. The early-life microbiome has also been associated with cognitive development in children. One study reported an inverse relationship between gut microbiome alpha diversity at 1 year of age and cognitive abilities at 2 years of age.Reference Carlson, Xia and Azcarate-Peril86 In a study of 309 children, gut microbiome composition at 3–6 months of age was associated with cognitive development at 3 years of age.Reference Sordillo, Korrick and Laranjo87 Many of the associations observed in this study were driven by taxa within the order Clostridiales.
These observations have been further explored in mechanistic animal studies. Diaz Heijtz et al. reported that germ-free mice showed altered motor activity and anxiety-like behaviour which was coupled with altered expression of genes involved in second messenger pathways and synaptic long-term potentiation in brain regions implicated in these behaviours.Reference Diaz Heijtz, Wang and Anuar6 Importantly, they could rescue these aberrant behaviours by conventionalising germ-free mice with normal gut microbiota in the gestational and neonatal period, but not adulthood. Germ-free mice also show impaired social behaviour and abnormal microglial development.Reference Forssberg88 Modulation of the maternal microbiome is sufficient to cause behavioural and neurological changes in the offspring in mice. Administration of non-absorbable antibiotics to pregnant mice reduces exploratory behaviour in their offspring.Reference Tochitani, Ikeno, Ito, Sakurai, Yamauchi and Matsuzaki89 Fostering these offspring with untreated dams from postnatal day 1 partially rescued this phenotype, suggesting that the maternal microbiome during gestation is involved in programming this behaviour. Stress induces changes to the gut microbiome. In mice, maternal prenatal stress alters both the maternal and offspring gut microbiota, and increases anxiety-like behaviour in the offspring.Reference Gur, Shay and Palkar90 These changes are accompanied by differences in cytokine expression in the placenta and the adult offspring amygdala. Modulation of the maternal microbiome using a HFD results in dysbiosis of the offspring microbiome coupled with altered social behaviour and neurological changes (specifically, changes in signalling in the mesolimbic reward system, oxytocin levels in the hypothalamus, and neural activity in the ventral tegmental area).Reference Buffington, Di Prisco, Auchtung, Ajami, Petrosino and Costa-Mattioli91 Importantly, this phenotype could be rescued after transferring stool from control mice to the offspring of HFD dams, demonstrating a causal link between maternal diet-induced dysbiosis, offspring dysbiosis, and the development of behavioural abnormalities. These authors were able to pinpoint a single bacterial species, Lactobacillus reuteri, which was drastically reduced in offspring of HFD dams. Treatment with L. reuteri restored social behaviour in these mice. The ability of L. reuteri to rescue social deficits in other models of autism spectrum disorder (including genetic, environmental, and idiopathic) has since been established.Reference Sgritta, Dooling and Buffington92 Together, these animal studies support observational evidence that the maternal and early-life microbiomes may influence neurological development and modulate the risk of neuropsychiatric disorders later in life.
Disruptions to the early-life microbiome
The human microbiome is vulnerable to disturbance in early life (Fig. 1). Dysbiosis of the maternal microbiome, intrapartum or neonatal antibiotic exposure, formula feeding, and delivery mode have all been shown to influence the early-life microbiome. Disturbances during this highly dynamic period appear to be transient in nature and are corrected after weaning.Reference Wampach, Heintz-Buschart and Hogan36,Reference Schloss, Iverson, Petrosino and Schloss93–Reference Fallani, Amarri and Uusijarvi95 Nevertheless, these events can have long-lasting impacts on the risk for later-life disease.
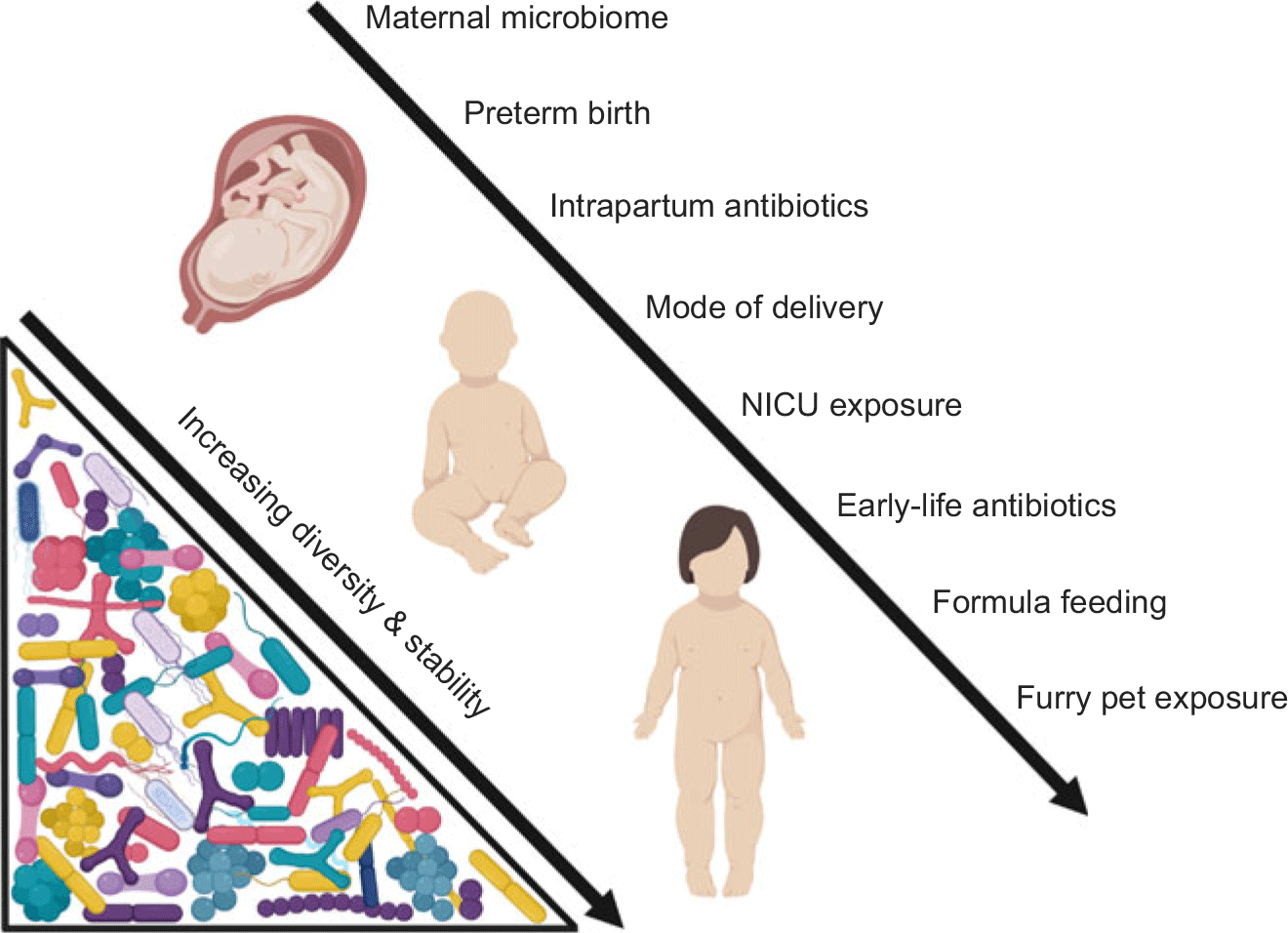
Fig. 1. Insults to the microbiome in early life (from gestation to early childhood).
Early-life antibiotics
Antibiotics are powerful disrupters of the gut microbiome across the lifespan; however, while the adult microbiome is generally able to recover from antibiotic exposure in a timely manner,Reference Rashid, Zaura and Buijs96 early-life antibiotics have longer-lasting effects. Antibiotics can have profound effects on the development of the early-life microbiome, although this is dependent on the type, dose, duration, and timing of administration.Reference Berardi, Pietrangiolillo and Bacchi Reggiani97 In one study of 142 Finnish children aged 2–7 years, early-life antibiotic use (from birth to the study period) was associated with long-lasting shifts in gut microbiota composition and metabolism.Reference Korpela, Salonen and Virta98 In this study, macrolides (drugs that inhibit bacterial protein biosynthesis, such as azithromycin) were shown to have a greater effect than β-lactams (drugs that inhibit bacterial cell wall biosynthesis, such as penicillin). In a randomised control trial, the effects of three commonly prescribed antibiotics were assessed in preschool-aged children.Reference Oldenburg, Sie and Coulibaly99 Children were randomised to receive amoxicillin, azithromycin, cotrimoxazole, or placebo for 5 days, with rectal swabs analysed before and after treatment. Azithromycin was found to decrease bacterial alpha diversity, while cotrimoxazole and amoxicillin had no discernible effect on diversity. Yassour et al. performed a longitudinal study of the gut microbiome of 39 children (half of whom were administered multiple course of antibiotics in the first 3 years of life) which included whole-genome shotgun sequencing of monthly stool samples, allowing strain-level resolution of the microbiome.Reference Yassour, Vatanen and Siljander100 Stool samples of antibiotic-exposed children had a lower level of bacterial diversity at both the species and the strain level. Further, the gut microbiome of antibiotic-treated children was less stable than that of untreated children.
Maternal antibiotic administration during pregnancy and birth has also been shown to disrupt the neonatal microbiome.Reference Nogacka, Salazar and Suarez101–Reference Fouhy, Guinane and Hussey106 Intrapartum antibiotic prophylaxis (IAP) has been shown to decrease bacterial diversity while decreasing the relative abundance of Bifidobacteria spp., Bacteroides spp., and Lactobacillus spp., and increasing the relative abundance of Enterococcus spp., and Clostridium spp.Reference Stinson, Payne and Keelan107 The mechanism by which maternally administered antibiotics affect the neonatal microbiome is unclear. Direct transmission of antibiotics via the placenta or breast milk may occur. Alternatively, antibiotics may cause temporary dysbiosis in the mother, resulting in transmitting her disrupted microbiota to her infant during the critical phases of early life. Perinatal antibiotic exposure has been associated with an increased risk of developing asthma,Reference Lapin, Piorkowski and Ownby108–Reference Stensballe, Simonsen, Jensen, Bonnelykke and Bisgaard111 allergies,Reference McKeever, Lewis, Smith and Hubbard112,Reference Wohl, Curry, Mauger, Miller and Tyrie113 and obesity.Reference Trasande, Blustein, Liu, Corwin, Cox and Blaser64–Reference Bailey, Forrest, Zhang, Richards, Livshits and DeRusso66,Reference Mueller, Whyatt and Hoepner69 These are important considerations, as administration of antibiotics during labour and delivery is a common practice. IAP is administered to all mothers delivering by caesarean section (CS; 34% of births in Australia114), all mothers who deliver vaginally and are vaginally or rectally colonised with group B streptococcus (20%–24% of births in AustraliaReference Angstetra, Ferguson and Giles115–Reference Hiller, McDonald, Darbyshire and Crowther117), as well as all mothers who experience prolonged rupture of membranes. Combined, these prophylactic interventions result may place a significant portion of the population at risk of early-life dysbiosis and associated later-life diseases.
Mode of delivery
Internationally, the rate of caesarean delivery has steadily increased between 2000 and 2015.114 In Australia, 34% of infants were born via CS in 2015.114 This is a significant concern as CS delivery has been repeatedly associated with early-life gut dysbiosis.Reference Wampach, Heintz-Buschart and Hogan36,Reference Backhed, Roswall and Peng38,Reference Azad, Konya and Maughan118–Reference Martin, Makino and Cetinyurek Yavuz122 CS-delivered infants are also at a greater risk of childhood asthma,Reference Debley, Smith, Redding and Critchlow123–Reference Thavagnanam, Fleming, Bromley, Shields and Cardwell125 atopic disease,Reference Laubereau, Filipiak-Pittroff and von Berg126–Reference Bager, Wohlfahrt and Westergaard128 allergies,Reference Bager, Wohlfahrt and Westergaard128,Reference Eggesbo, Botten, Stigum, Nafstad and Magnus129 obesity,Reference Darmasseelane, Hyde, Santhakumaran, Gale and Modi130 type 1 diabetes mellitus,Reference Cardwell, Stene and Joner131 inflammatory bowel disease,Reference Sevelsted, Stokholm, Bonnelykke and Bisgaard124,Reference Li, Tian and Zhu132 and impaired cognition.Reference Polidano, Zhu and Bornstein133 Some have ascribed this dysbiosis to a lack of exposure to the maternal vaginal microbiota at birth. Indeed, postnatal ‘vaginal seeding’ (i.e., swabbing of the maternal vaginal fluid across the skin and orifices of the newborn) has been trialled in an attempt to correct this supposed issue.Reference Dominguez-Bello, De Jesus-Laboy and Shen134 However, the act of passing through the vagina at birth may not be the factor that differentiates the microbiomes of CS-delivered and vaginally delivered infants. Instead, intrapartum antibiotic administration is likely a key factor in the observed CS microbiome.Reference Stinson, Payne and Keelan107 All CS-delivered infants are exposed to IAP, while the majority of vaginally delivered infants are not. Indeed, the gut dysbiosis caused by CS delivery closely mirrors that seen in vaginally delivered infants that are exposed to IAP, with a decrease in the abundance of Bacteroides and Bifidobacteria, and an increase in the abundance of Clostridium, Streptococcus, and Enterococcus.Reference Stinson, Payne and Keelan107 Further, women who deliver via CS tend to have higher BMIs,Reference Marchi, Berg, Dencker, Olander and Begley135–Reference Tun, Bridgman and Chari141 lower gestational age at delivery,Reference Delnord, Blondel and Drewniak142 and reduced rates of breastfeeding.Reference Dewey, Nommsen-Rivers, Heinig and Cohen143–Reference Evans, Evans, Royal, Esterman and James145 Regardless of the mechanism (IAP exposure or lack of vaginal microbiota exposure), mode of delivery is certainly associated with the microbiome in early life.
Preterm birth/NICU exposure
There is evidence to suggest that the human microbiome is seeded prior to birth.Reference Wampach, Heintz-Buschart and Hogan36,Reference Ferretti, Pasolli and Tett39,Reference Yassour, Jason and Hogstrom40,Reference Collado, Rautava, Aakko, Isolauri and Salminen146–Reference Jimenez, Marin and Martin149 However, this is a controversial topic and there is also evidence to suggest that bacterial exposure does not occur until birth.Reference Perez-Munoz, Arrieta, Ramer-Tait and Walter150 If humans are seeded with bacteria in utero, these are likely to play a significant role in the development of the fetal immune system.Reference Romano-Keeler and Weitkamp151 There is an increase in the rate of bacterial translocation across the intestines and an increase in trafficking of live bacteria around the body in the final trimester of pregnancy.Reference Perez, Dore and Leclerc152 This may suggest that bacterial exposure increases or perhaps commences in late pregnancy. Preterm birth may therefore bypass this prenatal exposure.
Additionally, preterm infants are subject to a range of medical interventions that have been associated with neonatal dysbiosis. In particular, preterm infants are exposed to the neonatal intensive care unit (NICU), the environment of which is likely to influence the neonatal microbiome.Reference Taft, Ambalavanan and Schibler153 These infants are also more likely to consume formula or donor breast milk (which is sterilised prior to use) and to experience less skin-to-skin contact with their mothers. Further, preterm infants are more likely to be delivered by CSReference Delnord, Blondel and Drewniak142 and to be administered antibiotics.Reference Patel, Konduru, Patra, Chandel and Panigrahi154 The sum of these aberrant early-life exposures is a significant difference in the neonatal gut microbiome of preterm infants compared to their full-term counterparts.Reference Ardissone, de la Cruz and Davis-Richardson155–Reference La Rosa, Warner and Zhou158
Breastfeeding
Breast milk contains a rich and diverse microbiome. Breastfeeding has been repeatedly shown to influence the development of the infant gut microbiome.Reference Stewart, Ajami and O’Brien35,Reference Azad, Konya and Persaud102,Reference Penders, Thijs and Vink119,Reference Thompson, Monteagudo-Mera, Cadenas, Lampl and Azcarate-Peril159–Reference Praveen, Jordan, Priami and Morine164 Exposure to breast milk bacteria and their metabolites modifies host gene expression and immune development.Reference Rogier, Frantz and Bruno162–Reference Praveen, Jordan, Priami and Morine164 Over a third of the gut bacteria of breastfed infants are vertically derived from their mothers’ breast milk and breast skin.Reference Pannaraj, Li and Cerini165 Additionally, breast milk contains prebiotics which modulate the infant gut microbiome and metabolome.Reference Marcobal and Sonnenburg166 A recent meta-analysis found that the gut microbiomes of exclusively breastfed infants persistently differed in composition, diversity, and function from non-exclusively breastfed infants.Reference Ho, Li and Lee-Sarwar167 In the TEDDY study, breastfeeding (exclusive or partial) was the most significant factor that shaped the gut microbiome in the first 4 years of life.Reference Stewart, Ajami and O’Brien35 In this study, there were many factors which significantly altered the infant gut microbiome at the genus and species level, but breastfeeding was the only factor that influenced the function of the microbiome. This is an important distinction, as it demonstrates a certain level of redundancy in the functional capacity of the early-life gut microbiome, which is considerably shaped by breastfeeding. The cessation of breastfeeding drives the maturation of the infant gut microbiome to an adult-like state.Reference Stewart, Ajami and O’Brien35,Reference Wampach, Heintz-Buschart and Hogan36
Given the important role of breastfeeding in the development of the infant microbiome, it is not surprising that formula feeding is associated with early-life dysbiosisReference Backhed, Roswall and Peng38,Reference Forbes, Azad and Vehling168–Reference Bezirtzoglou, Tsiotsias and Welling170 and the development of later-life NCDs.Reference Forbes, Azad and Vehling168,Reference Kelishadi and Farajian171,Reference Akobeng, Ramanan, Buchan and Heller172 However, the effect of expressing and bottle-feeding breast milk is less clear. Source-tracking studies have demonstrated that the skin around the areola is the source of approximately 10% of the breastfed infant microbiome.Reference Pannaraj, Li and Cerini165 Bottle-fed infants may therefore miss out on maternal skin microbes. Further, expressed breast milk is often stored frozen and thawed before use. The effect of these common practices on the infant gut microbiome is currently unknown, although there is evidence to suggest that hand-expressed milk varies in bacterial composition from pump-expressed milk.Reference Moossavi, Sepehri and Robertson173
Maternal microbiome
The maternal microbiome is the largest donor of bacteria to the infant microbiome.Reference Ferretti, Pasolli and Tett39,Reference Yassour, Jason and Hogstrom40 Therefore, maternal dysbiosis will negatively impact on the development of the infant microbiome. Factors that may skew the maternal microbiome include diet, stress, medications, dietary antimicrobials, diabetes, and obesity.
The gut microbiota of infants of obese mothers varies from that of infants of lean mothers,Reference Sugino, Paneth and Comstock174–Reference Collado, Isolauri, Laitinen and Salminen178 although this effect may be birth mode-dependent.Reference Mueller, Shin and Pizoni176 When stool from 2-week-old infants born of obese mothers (Inf-ObM) or infants born of lean mothers was transferred to germ-free mice, the Inf-ObM colonised mice showed gut dysbiosis, increased intestinal permeability, and increased susceptibility to obesity when exposed to a Western diet.Reference Soderborg, Clark and Mulligan179 These mice also displayed hallmarks of NAFLD. Importantly, all mothers in this study delivered vaginally, were not exposed to perinatal antibiotics, and exclusively breastfed. This suggests that maternal obesity is a causative factor in microbiome-mediated offspring obesity and NAFLD. Maternal transmission of an obesogenic microbiome may contribute to the intergenerational transmission of obesity. With approximately a third of all women of child-bearing age classified as obese,Reference Chen, Xu and Yan180 it is likely that a significant portion of the population is exposed to an obesogenic microbiome from birth. Further, obesity is a risk factor for other microbiome-modulating events, such as CS deliveryReference Chu, Kim and Schmid181 and reduced breastfeeding.Reference Lepe, Bacardi Gascon, Castaneda-Gonzalez, Perez Morales and Jimenez Cruz182
The maternal diet during gestation also shapes the offspring gut microbiome. In mice, 2-week-old pups of dams exposed to a Western diet had altered gut microbiota and sex-specific changes to colonic gene expression.Reference Steegenga, Mischke and Lute183 In a primate model, a perinatal maternal HFD, but not obesity per se, persistently altered the offspring gut microbiome.Reference Ma, Prince and Bader73 Offspring dysbiosis could not be fully corrected with a low-fat postnatal diet, suggesting that microbial exposures in utero and in early life programme long-term metabolism.
Perinatal stress can modulate the maternal and offspring microbiome and increase the risk of later-life disease. In a study of 56 vaginally delivered infants, maternal prenatal stress (both self-reported and inferred from serum cortisol levels) was associated with offspring gut dysbiosis, including a reduction in the relative abundance of Lactobacilli and Bifidobacteria, and an increase in the relative abundance of Escherichia, Serratia, and Enterobacter.Reference Zijlmans, Korpela, Riksen-Walraven, de Vos and de Weerth184 In a mouse model, maternal stress in early pregnancy disrupted the normal maternal microbiome adaptations to pregnancy (both gut and vaginal), resulting in temporal disruptions to the offspring microbiota.Reference Jasarevic, Howard, Misic, Beiting and Bale185 The effects of maternal stress on the offspring microbiota in mice have been implicated in altered hypothalamic pituitary adrenal (HPA) axis responses, immune dysfunction, and aberrant neural development, and can last long into adulthood.Reference Golubeva, Crampton and Desbonnet186,Reference Gur and Bailey187
Another major maternal condition which may detrimentally affect the early-life acquisition of microbes is diabetes. The intestinal and vaginal microbiota of women with gestational diabetes mellitus (GDM) varies from that of normoglycaemic pregnant women.Reference Ferrocino, Ponzo and Gambino188–Reference Wang, Zheng and Shi191 This maternal dysbiosis is mirrored in the infant gut microbiome.Reference Wang, Zheng and Shi191 Importantly, there is evidence to suggest that both GDM and pre-existing maternal diabetes mellitus are associated with changes in the amniotic fluid and meconium microbiomes.Reference Wang, Zheng and Shi191,Reference Hu, Nomura and Bashir192 These data might suggest that diabetes can alter the human microbiome prior to birth. One study, which analysed stool samples from mothers (n = 128) with and without GDM and their children (n = 109) 5 years after birth, found that the maternal microbiome had recovered at this time, while the microbiomes of children of GDM mothers remained altered.Reference Hasan, Aho and Pereira193 Given that gestational diabetes is a significant and increasing concern in the pregnant population, the impact of this disease on the early-life microbiome should be considered in more depth. In particular, the effect of diabetes and diabetic medication on the breast milk microbiome should be explored.
Household environment
Infants and young children spend the vast majority of their time inside the family home. The microbiome of the residential built environment is therefore an important consideration in the development of the early-life microbiome. Lynch et al. prospectively collected dust samples from homes with children <1 year old (n = 104) and measured atopy and wheeze in these children at 3 years of age.Reference Lynch, Wood and Boushey194 The household dust of children who did not develop atopy or wheeze by 3 years of age was richer and more diverse than that of affected children. These results suggest that household microbial exposures contribute to early-life immune development and may confer protection against common immune-mediated diseases.
The presence of furry petsReference Stewart, Ajami and O’Brien35,Reference Fujimura, Johnson and Ownby195,Reference Fujimura, Demoor and Rauch196 and older siblingsReference Stewart, Ajami and O’Brien35,Reference Laursen, Zachariassen and Bahl197 also has a significant effect on the early-life microbiome. Dust from homes with furry pets is microbially richer than that of homes without pets.Reference Fujimura, Johnson and Ownby195 There is a well-established link between pet ownership in early life and protection from allergy.Reference Hesselmar, Aberg, Aberg, Eriksson and Bjorksten198–Reference Ownby, Johnson and Peterson200 To test the hypothesis that this protection is conferred through exposure to pet-associated microbes, Fujimura et al. collected house dust from dog-owning and pet-free houses.Reference Fujimura, Demoor and Rauch196 Mice were exposed to these dust samples prior to and during an airway allergen challenge. Those animals sensitised with dog-residence dust had significant changes in the composition of their gut microbiomes and were protected from airway inflammation. This suggests that diversity within the environmental microbiome (particularly in cases of cohabitation with animals) confers a benefit to the developing microbiome.
Conclusions
The first years of life are a critical window for the development of the microbiome. Insults such as antibiotic use, maternal dysbiosis, and formula feeding during this time disrupt the early-life microbiome, increasing the risk for later-life metabolic, neuropsychiatric, and immunological disease. The prevalence of such microbiome-disrupting practices has increased over the past three decades and may have contributed to the rise of NCDs during this time. Importantly, the human microbiome is more readily changeable than the human gene content. Therefore, interventions to optimise the microbiome during gestation and infancy may help to lessen the societal burden of NCDs.
Acknowledgements
None.
Financial support
At the time of writing LFS was supported by a scholarship from the Women and Infants Research Foundation and The University of Western Australia.
Conflict of interest
None.
Ethical standards
None.



