INTRODUCTION
The HIV pandemic continues at a constant pace of about 15 000 new infections daily; viral diversity is a characteristic feature of the pandemic. HIV-1 subtype B is largely prevalent in Western Europe, North America and Australia, but it causes only 12% of infections worldwide while the greatest diversity is observed in developing countries where the majority of infections occur [Reference Geretti1–Reference Tatt3].
Non-B subtypes have spread throughout Europe in recent years [Reference Babic4–Reference Baldanti11]. Geographical distribution of viral lineages as well as significant pathways of virus dispersal across Europe has been described [Reference Paraskevis12]; however, this study focuses only on HIV-1 classified as subtype B throughout Europe, while current globalization leads to contacts between populations even outside the European Union, and subtype patterns may be heterogeneous in relation to diverse characteristics of the migration phenomenon in different areas. In this context, local surveillance of HIV epidemics is necessary.
We studied changes in prevalence of non-subtype B HIV-1 isolates across 3-year periods over more than 20 years using as denominator the number of new patients in each period. We also applied phylogenetic analysis to investigate possible transmission routes with the final aim of orientating targeted prevention strategies.
MATERIALS AND METHODS
Patients
A database was generated by merging the clinical data of the Institute of Infectious and Tropical Diseases and the HIV-1 genotypic data of the Institute of Microbiology of the University of Brescia. All patients who underwent HIV-1 drug resistance testing since the availability of the assay in 2000 were selected for this study. These patients were first diagnosed and entered HIV clinical care since 1983. The patients were ranked by calendar year of HIV diagnosis or first evaluation; therefore, prevalence of HIV-1 subtypes referred to the time when patients received HIV diagnosis or started follow-up rather than the time when resistance testing was performed. The time-frame of the study (1983–2006) was divided into 3-year periods. Temporal prevalences of HIV-1 subtypes were evaluated using as denominators patients who were diagnosed with HIV infection or entered clinical care in each period excluding patients who were diagnosed at earlier time-points. Prevalences were also evaluated according to geographic origin and risk factor for acquisition of HIV infection. Based on the self-reported risk factors, patients were characterized as intravenous drug users, homo-bisexuals, and heterosexuals. Subjects who denied any risk factor were classified as ‘other/unknown’.
HIV-1 sequence analyses
Peripheral blood was drawn by venepuncture into EDTA tubes, kept at room temperature, and centrifuged within 4 h of drawing at 1000 g for 15 min. Plasma was stored at −80°C and thawed in a 10–15°C water bath for 15 min before use. Specimens were tested for the presence of HIV RNA [Versant HIV-1 RNA 3.0 assay (bDNA); Bayer HealthCare, USA]. Samples were centrifuged at 23 500 g for 75 min at 4°C in a 2-ml microtube (PN 72.693.005; Sarstedt Inc., USA), and 800 μl of supernatant was removed. The manual extraction of 140 μl of original, diluted, or diluted-centrifuged samples was performed using the materials and Mini Spin protocol provided by the QIAamp viral RNA minikit (Qiagen Inc., USA) and absolute ethanol (catalogue no. E7023; Sigma-Aldrich, USA).
For HIV RNA sequencing, the manufacturer's directions for the TruGene HIV-1 genotyping kit and OpenGene DNA sequencing system were followed. The kit required diethyl pyrocarbonate-treated water (catalogue no. 9916; Ambion Inc., USA). Codons 4–99 of the protease gene and codons 38–247 of the reverse transcriptase gene of the pol region of the HIV-1 genome were amplified by PCR and sequenced.
Subtyping was obtained using the REGA HIV-subtyping tool (available at http://www.bioafrica.net/subtypetool/html/) associating the phylogenetic analysis and bootscan method. All sequences of non-subtype B and those supplied by REGA as unassigned were analysed for phylogenetic relationships. For this purpose, BiotEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) was used to align query sequences with pure subtype and circulating recombinant form (CRF) reference sequences obtained from the Los Alamos database (http://hivweb.lanl.gov/), and sequences were trimmed to obtain fragments of equal length (970 nt). The subtype assignment of the non-clade B strains was confirmed by a bootstrapped phylogenetic analysis using seqboot with 1000 replicates, followed by the DNAdist (with Kimura two-parameter method and a transition/transversion ratio of 2·0). Neighbor and Consense programs contained in PHYLIP (PHYLogeny Interference package (http://www.evolution.genetics.washington.edu/phylip.htlm). HIV-1 strains that did not cluster with reference subtypes or known CRFs were subsequently analysed by SimPlot software (version 2.5) (http://sray.med.som.jhmi.edu/RayAoft/Simplot/) both to calculate the percentage similarity between the query sequence and the number of reference of A–J subtypes, and to verify recombination in HIV-1 clades. The bootscanning method implemented in the SimPlot software was used to identify the subtypes involved in the recombination and their breakpoints. A neighbour-joining bootscan was also performed by comparing the query sequence with reference strains using a sliding window of 200 bp and moving forward in 20-bp steps.
Statistical analysis
To test the differences between subtypes B and non-B of the HIV-1 infected individuals the χ2 test was used for qualitative measures, while Student's t test was used to compare mean ages as appropriate. Cochran–Armitage test for trend was used to compare prevalence of HIV-1 subtypes across the 3-year periods. Univariate and multivariate logistic regression analyses were performed to investigate associations of variables (ethnicity, gender, route of transmission, calendar years) with non-subtype B infections. Two-sided P values <0·05 were considered to be statistically significant.
RESULTS
HIV-1 subtypes and general characteristics of patients
HIV-1 sequences were available from 1563 patients ranked by calendar year based on the date of the first positive HIV antibody test (84% of cases) or date of entry in the cohort where this information was missing (16%). The main characteristics of patients are illustrated in Table 1. According to REGA, 1237 (79·1%) sequences were classified as B subtype, 118 (7·5%) were pure non-B subtypes, 89 (5·6%) CRFs and 119 (7·6%) were unassigned. After bootscan and phylogenetic analysis, we identified additional 39 B subtypes (total=1276, 81·6%), three pure non-B subtypes (F1 subtypes) and 77 inter-subtype recombinants, 37 of which (31 from Italian, two each from European and Brazilian, and one each from African and Asian subjects) were either B/D or D/B inter-subtype recombinants and, as such, were considered B subtypes (http://www.hiv.lanl.gov/content/sequence/HIV/REVIEWS/LEITNER2005/leitner.html). Therefore, a total of 250/1563 (16%) non-subtype B viruses was attained. The sequences of these clinical isolates have been deposited in GenBank with the accession numbers GQ251042 to GQ251291.
Table 1. Patient characteristics and factors associated with non-subtype B infections (univariate and multivariate logistic regression analysis)
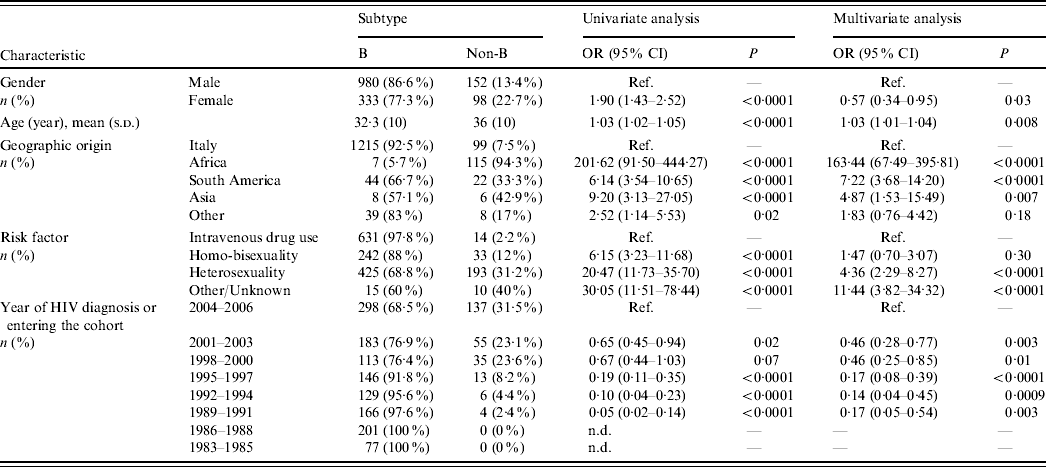
OR, Odds ratio; CI, confidence interval; Ref., reference category; n.d., not determined.
Odds ratios and 95% confidence intervals are not determined for periods 1983–1985 and 1986–1988 due to the presence of null values. Odds ratio for age is determined for each year.
Most Italian patients (92·5%) harboured HIV-1 B subtypes, while non-subtype B viruses were responsible for infection in 94% of subjects from Africa, 33% from South America and 43% from Asia (P<0·0001 for multiple group comparison across geographic origins including Italy). Multivariate logistic regression analysis demonstrated that risk of harbouring non-subtype B viruses was 163-fold, sevenfold and fivefold higher, respectively, in patients originating from these areas than in the Italian patients (Table 1). Notably, all patients from sub-Saharan Africa harboured non-subtype B viruses except for one who harboured B/D inter-subtype recombinant. In contrast, 33·4% of patients coming from South America harboured non-subtype B viruses (P<0·0001). Prevalence of HIV-1 non-subtype B viruses was higher in patients who were infected through heterosexual intercourse (31·2%) compared to intravenous drug users (2·2%) and homo-bisexuals (12%). Non-subtype B infections increased significantly over time (Cochran–Armitage trend test P<0·0001); prevalence of HIV-1 non-subtype B viruses in patients newly diagnosed or entering clinical care reached its peak level (31%) during 2004–2006. Logistic regression analysis confirmed the associations between non-subtype B infections and modes of HIV-1 transmission or calendar years, independently from geographic origin (Table 1).
Prevalence of HIV-1 subtypes in patients' categories over calendar years
Prevalence of non-subtype B HIV-1 viruses in subjects of Italian and non-Italian origin of those newly diagnosed or entering clinical care over time is shown in Figure 1 a. Prevalence of non-subtype B viruses increased over time not only because of an increasing number of non-Italian patients from endemic areas, but also because prevalence of Italian patients with HIV-1 non-subtype B infections increased significantly over time [from 0/76 (0%) in the period 1983–1985 to 2/129 (1·6%) in 1992–1994 and 67/331 (20·2%) in 2004–2006; Cochran–Armitage test for trend P<0·0001]. As shown in Figure 1 b, the proportion of patients with heterosexual intercourse as risk factor for HIV acquisition increased, reflecting the general trend of the HIV epidemic in Italy; percentage of HIV-1 non-subtype B viruses increased significantly over time in patients belonging to this risk category.

Fig. 1. HIV-1 B and non-B subtypes over time by (a) patients' nationality and (b) HIV transmission category.
HIV-1 subtypes and ethnicity
HIV-1 sequences from 70/99 (70·7%) Italian patients with non-subtype B infection clustered with pure non-B subtypes; 53 (53·5%) grouped with subtype F1, 12 (12%) with subtype C, three (3%) with subtype A1, and one (1%) each with subtypes D and G. Additional clinical isolates from eight (8%) and three (3%) patients, respectively, clustered with CRF02_AG and CRF01_AE while 18 (18%) patients were infected with inter-subtype recombinants (16 B/F, and two others). Table 2 shows the distribution of these mosaic forms according to geographic provenance.
Table 2. Distribution of inter-subtype recombinants according to patients' areas of origin (percentages are expressed in the total number of HIV-1 non-B subtypes found in patients from each area)
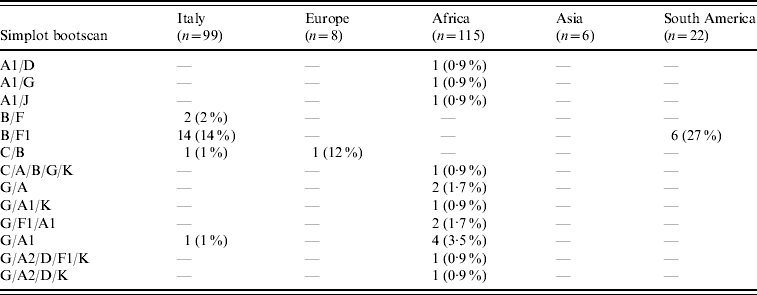
B/D or D/B inter-subtype recombinants were considered B subtypes (see http://www.hiv.lanl.gov/content/sequence/HIV/REVIEWS/LEITNER2005/leitner.html).
CRF02_AG was the most common subtype in African patients (67/122, 55%). Of African patients with non-subtype B viruses, CRF02_AG (67/115, 58·3%) was followed by subtype G (15/115, 13%), A1 and CRF06_cpx (six patients each), C (five patients) and CRF01_AE (one patient). Other inter-subtype recombinants were found in 15 patients (13%).
Distribution of HIV-1 non-B subtypes in South American patients resembled that of Italian patients. In fact, the most frequent non-B subtypes in patients from South America were F1 (9/22, 40·9%) and C (6/22, 27·3%). B/F inter-subtype recombinants were detected in six South American patients while CRF02_AG was present in only one patient. Interestingly, B/F inter-subtype recombinants appeared to increase both in Italian patients (none in patients diagnosed before 1995, one in 1995–1997, two in 1998–2000, three in 2001–2003, and 10 in 2004–2006) and in South Americans (none in patients diagnosed before 2001, three in 2001–2003 and three in 2004–2006). Last, subtype C (4/6) and A1 (3/6) were prevalent in Asian and non-Italian European subjects, respectively.
HIV-1 non-B subtypes by ethnicity and risk factors for HIV acquisition
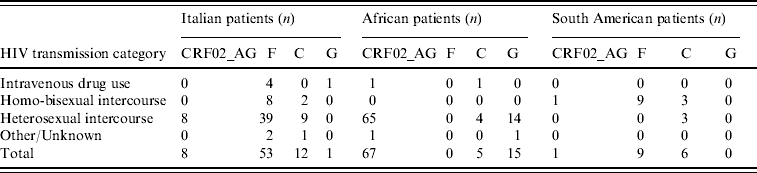
Table 3 illustrates the distribution of the most common non-subtype B viruses according to ethnicity and risk factors; only the most numerous ethnic groups are reported. The majority of Italian patients with F1 subtype reported heterosexual risk for HIV acquisition (39/53, 73·6%); 88% of these were males, and all but one (who had a history of travelling in Venezuela and Romania) denied risky behaviour for HIV acquisition in South America. All patients with F1 subtype from South America came from Brazil and were transsexuals.
Table 3. Distribution of the most frequent non-subtype B viruses or circulating recombinant forms in patients by HIV transmission category
Phylogenetic analysis
After assigning HIV-1 subtype, the phylogenetic relationships among viruses from patients with F1 or C subtypes were investigated with the aim of verifying eventual clusters of infections within these two groups which were the most numerous in our study population and also phylogenetically well-characterized compared to other subtypes, particularly inter-subtype recombinants (G, A, AG, BG, BF) [Reference Brindicci13–Reference Abecasis17]. Each phylogenetic tree (for C and F1 subtypes) included representative sequences downloaded from the Los Alamos database corresponding to isolates from different geographical areas in which the subtype is most commonly found. For example, sequences of subtype C from Brazil (n=4), India (n=7), Ethiopia (n=3), South Africa (n=4) and Botswana (n=4) were included, as well as subtype F1 sequences from Brazil (n=6) and Romania (n=4).
Figure 2 a shows the neighbour tree corresponding to subtype F1. In the study population, all strains but one clustered with representative sequences from Brazil and, overall, they apparently formed six different clusters (bootstrap >86%). The remaining sequence from an Italian patient (sequence 1496) segregated with representative strains from Romania.

Fig. 2. Phylogenetic relationships of pol sequences (protease, nt 2263–2560; reverse transcriptase, nt 2672–3302) from subjects infected with (a) subtype F1 and (b) subtype C. Different clusters are shown in red with the corresponding bootstrap value (1000 replicates).
The neighbour tree relative to subtype C, supported by a bootstrap value of >85%, is shown in Figure 2 b. Representative sequences from India, Brazil, Ethiopia and South Africa plus Botswana formed four separate country-specific clusters (monophyletic groups) indicating a founder-effect of the subtype C virus in these countries. Seven Italian strains, two African, one Asian and one strain originating from South America (sequences 1441–2, 1446, 1452, 1454–8, 1460, 1462) in our study population clustered together, as well as with representative sequences from Brazil (bootstrap value 64%). Moreover, within this segregation, five Italian, one South American and two African strains from our patients formed a group of tightly related sequences (bootstrap 88%) (1441–2, 1446, 1454–5, 1457, 1460, 1462). Two additional sequences (one Italian and one from an American descendant) clustered with the reference sequence C (BW, bootstrap value, 99%).
DISCUSSION
Our study is a contribution to the evaluation of HIV-1 non-subtype B virus prevalence and distribution in a large North Italian province, providing better knowledge of possible HIV transmission routes in order to target HIV prevention strategies.
Overall, 1563 patients were included in the study over a period of 23 years (1983–2006). The prevalence of non-subtype B viruses (16%) was the highest of those found in other Italian studies (5–13%) [Reference Geretti1]. This difference might be explained not only by different populations, but also by more recent estimates in the present study. Notably, prevalence of HIV-1 non-B subtypes peaked at 31% in HIV-positive patients diagnosed or entering clinical care in the latest 3-year period (2004–2006).
Even though immigration offers a valid explanation for the increasing prevalence of non-subtype B viruses in this area in which most migrants permanently reside (compared to other Italian areas where they are only passing through), the exponential increase of non-subtype B viruses among the Italian descendants (from 0% in 1983–1985 to 20% in 2004–2006) is worthy of note. Moreover, it is important to highlight the high prevalence of inter-subtype recombinants differing from currently accepted CRFs (http://hiv-web.lanl.gov/content/hiv-db/CRFs/CRFs.html), particularly in Italian descendants (18%).
This result which is due to the highly recombinogenic nature of HIV-1 has two main consequences; the first is epidemiological in that it reaffirms increasing circulation of multiple mosaic viruses in a previously clade B homogeneous area such as Italy [Reference Brindicci13]. The second consequence is methodological; in fact, according to REGA, 7·6% of sequences were unassigned, probably due to the fact that HIV-1 protease and part of the reverse transcriptase genes were examined. Since an accurate classification of recombinants is a prerequisite for epidemiological monitoring, on some occasions almost full-length genome analysis should be performed to provide reliable subtyping.
In our study, 37/1563 (2%) sequences, mostly from Italian, European and South American individuals, appeared as either B/D or D/B inter-subtype recombinants. A plausible explanation for this finding could be the common ancestral origin of B and D subtypes. In fact, for most genomic regions (especially env rather than pol gene), subtypes B and D should have been classified as sub-subtypes rather than separate subtypes because they are closer to each other than to other subtypes, but for the sake of consistency with previously published literature, subtypes B and D have not been renamed [Reference Paraskevis12, Reference Salemi18–Reference Worobey20].
Of particular interest was the identification of clusters of infections in the context of subtypes C and F1. We are aware that the methods used are somewhat imprecise and that results should be taken with caution; however, 1000 replications were performed on the neighbour-joining tree for the bootstrap analysis and only bootstrap values >85% were considered. Therefore, we are reasonably confident that a HIV transmission chain does exist, at least between South American and Italian descendants. This hypothesis is strengthened by the presence of B/F inter-subtype recombinants in South American and Italian patients, appearing in the two populations around 2000.
The overall prevalence of non-subtype B viruses in our population was higher in patients reporting heterosexual risk than in other risk groups, particularly in subjects of African and Italian origins. The finding that only 8/53 (15%) Italian patients infected with F1 subtype and 2/12 (17%) with C subtype reported homosexual intercourse, while all South American patients infected with F1 subtype and 3/6 with C subtype were homosexuals, would exclude a possible epidemiological link. However, this apparent contradiction might be due either to undisclosed (underreporting) homosexual contacts between Italian patients and transsexuals from Brazil or to established circulation of South American HIV-1 subtypes in the Italian population through heterosexual transmission. The acquisition of South American subtypes by Italian patients abroad could also be an explanation. The high prevalence of male patients who did not report foreign travel suggests that the first explanation is more reliable than the others.
In African patients, the most frequent non-B subtype was CRF02_AG. This recombinant form was also found in Italian patients, albeit at a frequency lower than that of South American subtypes and only in subjects who reported heterosexual risk. Thus, circulation of South American and African subtypes in the Italian population may be a consequence of different risk behaviours.
This study has some limitations. First, the patients were ranked by calendar year of HIV diagnosis or first evaluation; therefore, prevalence of HIV-1 subtypes referred to the time when patients received HIV diagnosis or started follow-up. The mixed approach used here may be prone to generate errors in the estimate of the prevalence over time. For example, latest years may have been characterized by a change in access to care for non-Europeans, resulting in an apparent, but false, increase in the prevalence of certain non-B subtypes. The fact that date of HIV diagnosis was available in most patients and the observation of increasing non-subtype B viruses in Italian patients seem to contradict this hypothesis. Second, in principle any assignment different from the actual year of infection is artificial. Thus, a cohort is suitable for analysis of the prevalence of the different clades over time if the cohort includes only patients with known seroconversion year. Unfortunately, the actual date of seroconversion was not available for most patients in our cohort. Third, the number of patients is still limited for assessment of temporal trends of every subtype indicating that our observations should be confirmed in larger cohorts. Fourth, due to the way HIV-1 sequences were generated (drug resistance testing), the analysis is limited to a fragment of the pol gene: a certain number of recombinants that would have been detected with analysis of a larger part of the genome may have been missed.
In conclusion, the HIV-1 epidemic is evolving in Italy with an increasing emergence of different non-subtype B viruses. Transmission of these strains is occurring mainly through sexual intercourse. The distribution of HIV-1 non-subtype B viruses in Italian patients, which mirrors that of South American patients, suggests a transmission chain of infection between these two ethnic groups, an established circulation of non-subtype B viruses within the Italian population, and the need for targeted prevention strategies.
ACKNOWLEDGEMENTS
This work has partially been funded by Virolab (www.virolab.org), which is sponsored by the European Commission (project IST-027446). We thank all colleagues who dedicated themselves to the care of patients and sampled analysis at the Institute of Infectious and Tropical Diseases and at the Virology Department of the University of Brescia, and the patients. We also thank Dr Laura Albini for revision and editing of the manuscript.
DECLARATION OF INTEREST
None.







