Mastitis is the most common disease in the bovine dairy industry. Many breeding programmes are affected while there is a lack of phenotypic data especially in mastitis selection where lowered somatic cell count (SCC) is taken as a surrogate of clinical mastitis (Rupp & Boichard, Reference Rupp and Boichard1999). Single nucleotide polymorphisms (SNPs) within genes of the local mammary innate immune response are attracting particular importance as potential DNA markers for mastitis susceptibility (Chen et al. Reference Chen, Yang, Ji, Mao, Chen, Li, Wu, Wang and Chang2011). Mitogen activated protein kinase kinase kinase kinase (MAP4K4) is a member of the germinal centre kinase group related to Saccharomyces cerevisiae (Zohn et al. Reference Zohn, Li, Skolnik, Anderson, Han and Niswander2006; Delpire, Reference Delpire2009). Many experiments have been conducted to link the expression and functions of MAP4K4 to various patho-physiological processes including metabolism, inflammation, neural degeneration and cancer. It has been suggested that inhibition of MAP4K4 function may be beneficial in treating disease associated with these processes. MAP4K4 gene is responsible for the key processes in initiation of innate response (Dror et al. Reference Dror, Alter-Koltunoff, Azriel, Amariglio, Jacob-Hirsch, Zeligson, Morgenstern, Tamura, Hauser, Rechavi, Ozato and Levi2007). Since MAP4K4 is an important gene that regulates inflammation, it would be of use to study the role of MAP4K4 in mastitis resistance. MAP4K4 is a good candidate gene since together with IL-17R and SOCS7 it plays a key role in the NF-kB signalling pathway directing transcriptional regulation of pro-inflammatory genes in macrophages (Karin & Greten, Reference Karin and Greten2005; Dror et al. Reference Dror, Alter-Koltunoff, Azriel, Amariglio, Jacob-Hirsch, Zeligson, Morgenstern, Tamura, Hauser, Rechavi, Ozato and Levi2007). Identification of SNPs in bovine MAP4K4 could yield useful information (Schulman et al. Reference Schulman, Sahana, Iso-Touru, Lund, Andersson-Eklund, Viitala, Varv, Viinalass and Vilkki2009) and we hypothesised that MAP4K4 gene could have an important role in immunity and could be associated with mastitis resistance. To test this hypothesis, an association study investigating the relationship between bovine MAP4K4 polymorphism and mastitis and different milk traits in dairy cows was performed.
Material and methods
All experiments were carried out according to the regulations for the Administration of Affairs Concerning Experimental Animals published by the Ministry of Science and Technology, China in 2004 and approved by the Animal Care and Use Committee from the Dairy Cattle Research Center, Hubei Academy of Agricultural Sciences, Hubei, P. R. China.
Animal management and blood sampling
The dairy herd, located in Hubei province, China, belongs to the sub-humid tropical climates, where the annual average temperature is 16–17 °C and the annual rainfalls is 1100–1200 mm with 75% relative humidity. Cows were housed in free stall barns and fed with total mixed ration (TMR) thrice daily. 10 ml Blood samples from 500 Chinese Holstein cows with good DHI were collected in a tube with EDTA (pH = 8.0) anticoagulants and samples were kept in −20 °C until the genomic DNA was extracted. DNA was extracted according to the method described by Wang et al. (Reference Wang, Liu, Li, Ju, Huang, Li, Wang and Zhong2011). After genomic DNA isolation, the quantity and purity of DNA were measured using NanoDrop™ ND-2000c Spectrophotometer (Thermo Scientific, Inc.)
Primer design, sequencing and genotyping
Information from Ensembl Genomic Browerser (http://www.ensembl.org), sequence of MAP4K4 gene referring to Bos_taurus_UMD_3·1 assembly (ENSBTAG00000013023) exonic and intronic region of MAP4K4 gene was annotated. We designed 34 pairs of primers (Supplementary File S1) to amplify 5′ upstream region, 3′ UTR, 3′ downstream region, exonic and adjacent intronic regions of bovine MAP4K4 gene. All the primers were designed using the Primer Premier 5·0 software (Premier Biosoft International, Palo Alto, CA, USA) and synthesised by Sangon Biological Engineering Technology (Shanghai, China). Briefly, the total 25 µl volume of mixture contained 50 ng genomic DNA, 12·5 µl 2X reaction mix (including 500μM dNTP each; 20 mM Tris–HCl; pH 9; 100 mM KCl; 3 mM MgCl2), 0·5 µM of each primer, and 0·5 units of Taq DNA polymerase. The cycling protocol was as follows: 5 min at 95 °C, 35 cycles of denaturing at 94 °C for 30 s, annealing at T m°C for 30 s, extending at 72 °C for 30 s, with a final extension at 72 °C for 10 min.
The PCR products with different electrophoretic patterns were directly sequenced in both directions using an ABI PRISM 3730 DNA analyser (Applied Biosystems, Foster City, CA, USA) following the standard protocol. The DNAMAN software package (Version 6.0; Lynnon Biosoft, Quebec, Canada) was used to analyse the sequencing results and to determine the mutation position. Selection of enzymes for genotyping was done with software http://watcut.uwaterloo.ca/template.php. The PCR products were digested with the corresponding restriction endonucleases (Supplementary File S3) at 37 °C for 8 h, according to manufacturer recommendations. The digested products were observed on a 2·5% agarose gel stained with ethidium bromide and final genotype of the animal was determined.
Statistical analysis
Exact test of Hardy–Weinberg Equilibrium was conducted before the association study using the R package ‘genetics’ SNPs were filtered for deviation from Hardy-Weinberg Equilibrium (P-value ≤ 0·05). The associations between the retained SNPs and the traits (fat percentage, protein percentage, SCS, 305 d milk yield) were analysed using the general linear model (GLM) procedure of R. The general linear model is as following:
Where y ijk is the observed value, μ is the overall population mean, F i is the effect of the ith farm, P j is the effect of the jth parity, SNP k is the effect of the kth single SNP. e ijk is the random error. Tukey Honest Significant Differences (Tukey HSD) method was used for multiple comparison between different genotypes.
Results and Discussion
Characterisation of boMAP4K4 SNPs and genotype allelic frequencies
A total of 19 SNPs were detected in theMAP4K4 gene(ENSBTAG00000013023) of which 4 were located in 5′upstream region, eight in exons and seven in introns. Seven SNPs were synonymous substitutions and one was non-synonymous substitution (c.1562C/T) that changed amino acid Ser into Phe (Supplementary File S2). Analysis of this 1200 bp sequence by UCSC/Ensembl showed the region in chr11: 6609650-11:6609699 where it was marked as a promoter. Similarly, we identified no miRNA binding sites at 3′UTR(ENSBTAG00000013023) MAP4K4 gene. Analysis of these results with identified SNPs results revealed that none of the SNPs were found within these potential regions. Nine SNPs were selected for the genotyping, of which eight were within Hardy–Weinberg equilibrium (Supplementary Files S3 and S4). The 9th SNP (c.2115A/G) did not fit with Hardy–Weinberg equilibrium (P < 0·01).
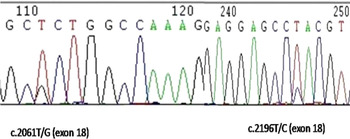
Fig. 1. Sequencing for the identified SNPs at exon 18 (c.2061T/G and c.2196T/C) of MAP4K4 gene on cow chromosome 11.
Analysis of boMAP4K4 SNPs with milk traits and SCS
We calculated the somatic cell score from somatic cell count to analyse the probable resistibility/ susceptibility of animals for mastitis. The analysis revealed SNP3 (C.2061T/G) and SNP4 (C.2196T/C) to be significant for somatic cell scores (Figure 1). In both these SNPs the homozygote (TT) animals were found to have significantly higher (P < 0·01) SCS than heterozygote (TG) and (TC) animals respectively (Table 1). Analysis of productivity traits within the genotyped animals revealed that the SNP6 (11:6646091G/A) and SNP8 (11:6646308A/G) at intron 21 were significantly associated with fat percentage. In SNP6, the AA genotype animals had significantly (P < 0·05) higher fat percentage when compared with the heterozygote GA and homozygote GG had animals. A similar pattern was observed in SNP8, where the homozygote GG had significantly higher (P < 0·01) fat percentage compared with the homozygote CC (Table 1).
Table 1. Analysis of boMAP4K4 SNP genotype by SCC and milk productivity trail
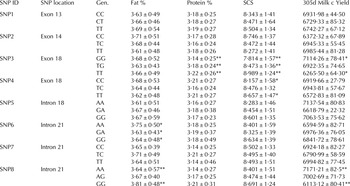
* P < 0·05
** P < 0·01
The MAP4K4 gene plays a crucial role in the innate and acquired immune systems and a number of studies in humans showed that its genetic variation may alter the body's susceptibility to infectious diseases (Bouzakri et al. Reference Bouzakri, Ribaux and Halban2009). There have not been any reports concerning the characterisation of MAP4K4 gene in cow genome and the identification of SNPs associated with milk traits. Results from human and other species shows that MAP4K4 is a strong candidate gene for resistance to bacterial diseases in mastitis (Karin & Greten, Reference Karin and Greten2005; Dror et al. Reference Dror, Alter-Koltunoff, Azriel, Amariglio, Jacob-Hirsch, Zeligson, Morgenstern, Tamura, Hauser, Rechavi, Ozato and Levi2007). It is important to screen the polymorphisms inMAP4K4 gene and elucidate its effect on immune cell functions which would help to facilitate breeding aimed at disease-resistance by marker-assisted selection method in mastitis. Analysis of bovine MAP4K4 SNPs may contribute to the discovery of mastitis resistance related genetics traits because bacteria are the most common cause of mastitis in cattle. The SCS has been used as the criterion for improving mastitis resistance (Shook & Schutz, Reference Shook and Schutz1994). Statistical analysis revealed that cows with c.2061/G -GG and c.2061/G-TG genotypes had significantly lower SCSs than the TT subjects, whereas the cows with c.2196C/T-CC had significantly lower SCSs than the TT subjects, which indicates that genotypes GG, TG and CC might be associated with mastitis resistance. Allele T of SNPs c.2061/G and T of SNP c.2196C/T may predispose cows to clinical mastitis via increased neutrophil trafficking to mammary gland, because clinical mastitis is correlated with SCS, as reported by Rupp & Boichard (Reference Rupp and Boichard1999). Thus, cattle with genotypes GG, TA and CC could be used for breeding strategy.
In conclusion, our findings confirm that MAP4K4 c.2061/G and c.2196C/T could be regarded as novel candidate genetic molecular markers for mastitis resistance/susceptibility in Chinese Holstein. However, this does not imply that genetic selection for mastitis resistance should be based on only Bo-MAP4K4 alleles. The possible associations of c.2061/G and c.2196C/T with mastitis resistance and the occurrence of preferable haplotypes within the MAP4K4 gene are interesting topics for future studies.
The authors are grateful for financial assistance from the research platform of the Ministry of Agriculture and International Exchange and Cooperation in Science and Technology and EUFP7 projects (PIIFR-GA-2012-912205 and FP7-KBBE-2013-7-613689).
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0022029916000832




