Feed for intensively farmed fish still relies heavily on feedstuffs of marine origin, fishmeal and fish oil. This impairs the sustainability of fish production, while aquaculture should be a solution to the generally observed decline in fishery resources(1). Research is intense for finding ways to replace marine feedstuffs (fishmeal and fish oil) by plant feedstuffs(Reference Gatlin, Barrows and Brown2). In the past 20 years, fish feeds have included large amounts of fish oil, given the beneficial effects on N utilisation and environmental load(Reference Cho and Kaushik3, Reference Sargent and Tacon4). Partial and total replacement of fishmeal by vegetable protein sources is similarly the object of several studies in almost all species(Reference Gatlin, Barrows and Brown2, Reference Webster and Lim5).
Efforts towards replacement of fishmeal by other alternative protein sources have been undertaken for more than two decades and there is a vast amount of literature on partial replacement of fishmeal by plant feedstuffs(Reference Webster, Lim and Lee6). A number of disadvantages have been ascribed to the use of plant protein sources: relatively low protein content, amino acid imbalance, low palatability, presence of endogenous anti-nutritional factors and large amounts of carbohydrates(Reference Gatlin, Barrows and Brown2, Reference Francis, Makkar and Becker7, Reference Kaushik, Flos, Tort and Torres8). Attempts have been made to develop fishmeal-free diets for different species including salmonids either with single ingredients duly supplemented with amino acids(Reference Kaushik, Cravedi, Lalles, Sumpter, Fauconneau and Laroche9) or using a mixture of different protein sources(Reference Watanabe, Aoki, Shimamoto, Hadzuma, Maita, Yamagata, Viswanath and Satoh10–Reference Gomez-Requeni, Mingarro, Calduch-Giner, Medale, Martin, Houlihan, Kaushik and Perez-Sanchez13). It is clear that a substantial reduction in the dietary levels of fishmeal can be achieved although total replacement of fishmeal by plant ingredients is still not common in salmonids. Some earlier studies have shown that total replacement of fishmeal by plant proteins leads to decreased growth of rainbow trout possibly linked to a modification of a number of hepatic metabolic pathways(Reference Vilhelmsson, Martin, Medale, Kaushik and Houlihan14).
Several studies with salmonids (rainbow trout, brown trout, Atlantic salmon, Pacific salmons) have shown that it is possible to replace fish oil by a single vegetable oil or mixture of vegetable oils without affecting growth or feed efficiencies(Reference Sargent and Tacon4, Reference Bell, Tocher, Henderson, Dick and Crampton15). Since the flesh fatty acid composition is known to be affected by the dietary fatty acid profiles, it is also known that once the fish are grown with vegetable oils over the major part of the life cycle, a finishing diet based on fish oil as the major lipid source can be used to tailor the final flesh fatty acid composition with the levels of n-3 PUFA (EPA and DHA) ideally suited for human nutrition and health(Reference Bell, Tocher, Henderson, Dick and Crampton15–Reference Regost, Arzel, Robin, Rosenlund and Kaushik17). The metabolic consequences are also numerous, mediated by a number of interacting pathways.
The objective of the present study was to analyse the hepatic gene expression profile in rainbow trout (Oncorhynchus mykiss) fed over a long period diets with or without either fish oil or fishmeal, replaced respectively by a mixture of plant oils or plant proteins. We analysed specifically the liver since this is the main organ involved in nutrient utilisation as the centre of intermediary metabolism in animals. After transcriptomic analysis, differentially expressed genes were identified and some were specifically studied in rainbow trout liver following the plant-based diet intake.
Experimental methods
Feeds, fish rearing and sampling
Triploid rainbow trout were reared in the French National Institute for Agricultural Research (INRA) experimental fish farm at constant water temperature (17 ± 1°C) and under natural photoperiod conditions (Donzacq, Landes, France). To test the effect of fish oil replacement, fish were fed from first feeding to commercial size during 62 weeks with two isoproteic (51 % crude protein), isolipidic (30 % crude fat) and isoenergetic (26 kJ/g) diets, differing only by the lipid source, i.e. either fish oil or a mixture of vegetable oils (30 % palm, 15 % linseed, 55 % rapeseed) as previously described(Reference Richard, Kaushik, Larroquet, Panserat and Corraze18) (Table 1).
Table 1 Composition of the diets used in the fish oil replacement studies*

FO diet, fish oil diet; VO, vegetable oil diet; CP, crude protein.
* For further details, see Richard et al. (Reference Richard, Kaushik, Larroquet, Panserat and Corraze18). Diets were produced at Nutreco Technology Centre, Norway as extruded pellets.
† Mineral and vitamin premix according to National Research Council recommendations.
To test the effect of fishmeal replacement, fish were fed from first feeding to commercial size during 52 weeks with two isonitrogenous, isolipidic and isoenergetic diets, differing only by the protein source, i.e. either fishmeal or a mixture of plant proteins (Table 2). These diets were produced by feed manufacturers (Nutreco, Stavanger, Norway and Le Gouessant Aquaculture, Lamballe, France, respectively).
Table 2 Composition of the diets used in the fishmeal replacement studies*

FM diet, fishmeal diet; PP diet, plant-protein diet; CP, crude protein.
* Diets were produced at Le Gouessant Aquaculture, France as extruded pellets.
† Mineral and vitamin premix according to National Research Council recommendations.
Fish were randomly distributed into triplicate tanks per dietary treatment. Each diet was distributed by hand to visual satiation 6 d over 7 d and feed consumption was recorded every week. At the end of the growth trial, six fish from each group (two per tank) were randomly sampled 24 h after the last meal in order to have data following the long-term plant-diet adaptation. Fish were killed by a sharp blow to the head. Livers were weighed and immediately frozen in liquid N2 and kept at − 80°C pending analyses.
Chemical composition of the diets
The experimental diets were analysed using the following procedures. DM was determined after drying at 105°C for 24 h. Gross energy was determined using an adiabatic bomb calorimeter (IKA; Heitersheim Gribheimer, Germany). Protein content (N × 6·25) was determined by the Kjeldahl method after acid digestion. Total lipid content was determined by the method of Folch et al. (Reference Folch, Lees and Sloane Stanley19), after extraction by dichloromethane rather than chloroform. Fatty acid composition of the diets (and the whole body of fish fed with or without fish oil) was determined in the total lipid extract after acid-catalysed transmethylation as previously described(Reference Richard, Kaushik, Larroquet, Panserat and Corraze18). Amino acid composition was determined after acid hydrolysis: amino acids were separated by ion-exchange chromatography using pH gradient elution followed by post-column derivatisation ninhydrin according to the method of Moore & Stein(Reference Moore and Stein20).
cDNA microarrays
Nylon microarrays were obtained from INRA-GADIE biological resources centre (Jouy-en-Josas, France; http://www-crb.jouy.inra.fr/). A total of 9023 rainbow trout cDNA originating from a pooled-tissue library(Reference Govoroun, Le Gac and Guiguen21) plus 193 controls were spotted after PCR amplification. PCR products were spotted onto Hybond N+ membranes as described by Nguyen et al. (Reference Nguyen, Rocha, Granjeaud, Baldif, Bernard, Naquet and Jordan22). Positive (plant luciferase cDNA depot) and negative (water depot) controls were also spotted on each microarray.
Hybridisation, scanning and quantification of microarrays
Total RNA were extracted from rainbow trout liver using TRIzol® reagent (Invitrogen, Carlsbad, CA, USA). Four hepatic RNA samples corresponding to four individuals per dietary group were used for microarray hybridisation at INRA UMR1067 transcriptomic facility (St-Pée-sur-Nivelle, France) according to the following procedure. RNA quality was determined using an Agilent bioanalyser. A first hybridisation was performed at 42°C for 48 h using γ33P-labelled T7 promoter oligonucleotide (5′-CACTATAGGGAATTTGGCC-3′) to estimate the amount of cDNA in each spot. After stripping (3 h at 68°C, 0.1X SSC, 0·2 % SDS), hybridisations with hepatic cDNA were performed. Microarrays were prehybridised for 1 h at 65°C in hybridisation buffer (5X Denhardt, 5X SSC, 0·5 % SDS). Labelled cDNA were prepared from 5 μg RNA by simultaneous reverse transcription and labelling for 1 h at 42°C in the presence of 1·85 MBq (50 μCi) [α- 33P] dCTP, 5 μm-cold dCTP and 800 μm each of dATP, dGTP and dTTP and 200 units SuperScript™ III RT (Invitrogen) in 30 μl final volume. A positive control corresponding to the luciferase mRNA (20 ng) (Promega, Madison, WI, USA) was simultaneously prepared. RNA was degraded by treatment at 68°C for 30 min with 10 % SDS (1 μl), 0·5 m-EDTA (1 μl) and 3 m-NaOH (3 μl) and then equilibrated at room temperature for 15 min. Neutralisation was done by adding 1 m-2-amino-2-hydroxymethyl-propane-1,3-diol-HCl (10 μl) and 2 m-HCl (3 μl). Microarrays were then incubated with the corresponding denatured labelled cDNA for 48 h at 65°C in hybridisation solution. After three washes (1 h at 68°C with 0.1X SSC, 0·2 % SDS), microarrays were exposed for 65 h to phosphor-imaging plates that were scanned using a Fuji BAS-5000 (Fuji, Tokyo, Japan). Signal intensities were quantified using AGScan software (bioinformatic platform Sigenae; INRA; http://www.sigenae.org/)(Reference Lopez, Rougemont, Loriod, Bourgeois, Loi, Bertucci, Hingamp, Houlgatte and Granjeaud23, Reference Cathelin, Lopez and Klopp24).
Microarray data analysis
Data microarrays were deposited in BioArray Software Environment (BASE) database(Reference Saal, Troein, Vallon-Christersson, Gruvberger, Borg and Peterson25), a ‘minimum information about a microarray experiment’ (MIAME) supportive customisable database available at the bioinformatic platform Sigenae. Signal processing was performed using vector oligonucleotide data to correct the relative amount of DNA present in each spot. At this step, low nucleotide signals (less than three times the background level) were excluded from the analysis. After correction, the signal was normalised by dividing each gene expression by the median value of the array before log transformation. Data were subsequently analysed using statistical TIGR Multiple Experiment Viewer software (TMEV; The Institute for Genomic Research, J. Craig Venter Institute, Rockville, MD, USA), which is a suite of microarray data analysis applications. Variation of gene expressions between two dietary treatments was termed significant when the P value was < 0·01 using the two-sample t test for microarrays(Reference Pan26) and followed by supervised hierarchical clustering for significant genes only. Organisation of genes for biological interpretation in the context of gene ontology was performed using GoMiner software (Genomics and Bioinformatics Group, National Institutes of Health, Bethesda, MD, USA; http://discover.nci.nih.gov/gominer/)(Reference Zeeberg, Feng and Wang27).
Data mining
Rainbow trout sequences originating from INRA Agenae(Reference Govoroun, Le Gac and Guiguen21) and the US Department of Agriculture(Reference Rexroad, Lee, Keele, Karamycheva, Brown, Koop, Gahr, Palti and Quackenbush28) and expressed sequence tag (EST) sequencing programs were used to generate publicly available contigs (http://public-contigbrowser.sigenae.org:9090/index.html). The 4th version (om.4) was used for BlastX (version 4 (om.4); Basic Local Alignment Search Tool; http://www.ncbi.nlm.nih.gov/blast/) comparison. The score of each alignment was retrieved after performing a BlastX comparison.
Real-time RT-PCR
Gene expression levels were determined by real-time RT-PCR using six RNA including those used for microarray analysis. Total RNA (1 μg) was reverse transcribed to cDNA with the Superscript™ III RNAse H RT kit (Invitrogen, Carlsbad, CA, USA) using oligo dT primers. Real-time PCR was performed in the iCycler iQ™ (Bio-Rad, Hercules, CA, USA). Quantitative PCR analyses for gene expressions were performed on 10 μl of the RT reaction mixture using the iQ™ SYBR® Green Supermix (Bio-Rad). The total volume of the PCR reaction was 25 μl containing 200 nm of each primer. Primers were designed so that they were overlapping an intron when it was possible (Primer3 software; http://biotools.umassmed.edu/bioapps/primer3_www.cgi) using known sequences in nucleotide databases (Table 3).
Table 3 Primer pairs for analysis by real-time PCR of selected genes*

* Nucleotide sequences were extracted from the Sigenae database (http://www.sigenae.org/) except for GS01, GS02 and GS03 from Genbank(Reference Essex-Fraser, Steele, Bernier, Murray, Stevens and Wright45).
† Reverse transcription with random primers.
Thermal cycling was initiated with the incubation at 95°C for 90 s for hot-start iTaq™ DNA polymerase activation. Thirty-five steps of PCR were performed, each one consisting of heating at 95°C for 20 s for denaturing, and at 59°C for 30 s for annealing and extension. Following the final cycle of the PCR, melting curves were systematically monitored (with a gradient of 0·5°C per 10 s from 55 to 94°C) to ensure that only one fragment was amplified. Samples without RT and samples without RNA were run for each reaction as negative controls.
Data analysis
Data are presented as mean values and standard deviations. We analysed the effects of the different diets with an unpaired two-tailed Student's t test (Systat 9 software products; SPSS, Inc., Chicago, IL, USA), except for microarray data (see before) and quantitative RT-PCR data. For the latter, significant differences were considered at P < 0·05. Relative quantification of the target gene transcript with the ef1α reference gene transcript(Reference Olsvik, Lie, Jordal, Nilsen and Hordvik29) was made following the Pfaffl method with the Relative Expression Software tool (REST©)(Reference Pfaffl30, Reference Pfaffl, Horgan and Dempfle31). This mathematical algorithm computes an expression ratio, based on real-time PCR efficiency and the crossing point deviation of the unknown sample v. a control group:

where E is PCR efficiency determined by standard curves using serial dilution of cDNA (cDNA dilutions from 1/16 up to 1/512), ΔCT being the crossing point deviation of an unknown sample v. a control. Statistical differences in gene expression between control and sample were evaluated in group means by randomisation tests(Reference Pfaffl, Horgan and Dempfle31) using REST© software. A total of 2000 random allocations were performed and significant differences were considered at P < 0·05.
Results
Fish rearing and endpoint analysis: growth rate, feed efficiencies and whole-body composition
In the fish fed with fish oil and vegetable oils, growth performance was similar throughout the long-term study irrespective of dietary treatment; at the end of the growth study the two groups reached a final body weight of about 1 kg (see Richard et al. (Reference Richard, Kaushik, Larroquet, Panserat and Corraze18) for further details). No differences were found in feed efficiency, protein feed efficiency and feed intake (Table 4). As it is well known that replacement of fish oil by vegetable oils may produce major changes in fatty acid composition of fish, we also analysed the whole-body fatty acid composition for the fish oil- and vegetable oil-fed fishes (Table 5); the whole-body fatty acid composition reflected largely the composition of the diet (Table 1). The fish fed vegetable oils exhibited the highest levels of 18 : 1, 18 : 2n-6 and 18 : 3n-3, whereas the fish fed the fish oil diet had a very high percentage of long-chain MUFA (14·1 % for 20 : 1 and 6·9 % for 22 : 1) and also the highest proportion of EPA and DHA (2·4 and 5·2 % respectively). In contrast to the fish fed with vegetable oils, trout fed with plant proteins had significantly lower growth rates, lower feed efficiency and lower protein feed efficiency than fish fed with fishmeal (P < 0·05; Student's t test) (Table 6), even though the feed intake was higher in the fish fed vegetable proteins. These data suggest a low capacity of metabolic adaptation of the fish fed with plant proteins from first feeding.
Table 4 Effects of fish oil replacement on growth performance and feed efficiency in juvenile rainbow trout over 62 weeks (initial body weight (IBW) 0·120 g)*
(Mean values and standard deviations for three tanks)
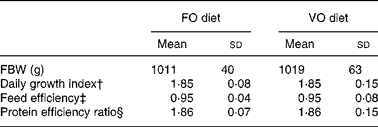
FO diet, fish oil diet; VO, vegetable oil diet; FBW, final body weight.
* There was no significant difference between the groups (P >0·05; Student's t test).
† Daily growth index = 100 × (FBW1/3 − IBW1/3)/duration (49 d).
‡ Feed efficiency = wet weight gain (g)/dry feed intake (g).
§ Protein efficiency ratio = wet weight gain (g)/crude protein intake (g).
Table 5 Effect of fish oil replacement on mean fatty acid composition of whole-body lipids after feeding the experimental diets for 62 weeks
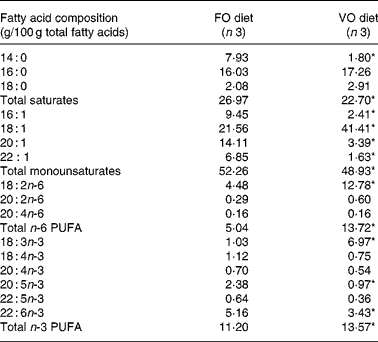
FO diet, fish oil diet; VO, vegetable oil diet.
* Mean value was significantly different from that of the fish fed the FO diet (P < 0·05).
Table 6 Effects of fishmeal replacement on growth performance and feed efficiency in juvenile rainbow trout over 52 weeks (initial body weight (IBW) 0·215 g)
(Mean values and standard deviations for three tanks)
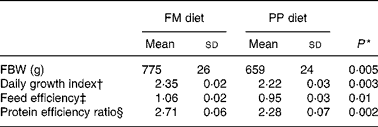
FM diet, fishmeal diet; PP diet, plant-protein diet; FBW, final body weight.
* By Student's t test.
† Daily growth index = 100 × (FBW1/3 − IBW1/3)/duration (49 d).
‡ Feed efficiency = wet weight gain (g)/dry feed intake (g).
§ Protein efficiency ratio = wet weight gain (g)/crude protein intake (g).
Differentially expressed genes in the liver of rainbow trout linked to diet composition
Analysis of microarray data showed that almost 0·8 % of genes were differentially expressed in our two experimental comparisons: (i) between the fish fed fish oil and vegetable oils, and (ii) between the fish fed fishmeal and plant proteins (Tables 7–10) (P < 0·01, t test; TMEV). Among the seventy-one genes differentially expressed between the fish fed with fish oil and those fed with vegetable oils, sixteen and fifty-five hepatic transcripts exhibited, in the fish fed with vegetable oils, higher and lower abundance respectively (Tables 7 and 8). Moreover, among the seventy-five genes differentially expressed between the fish fed with fishmeal and those fed with plant protein, fifteen and sixty hepatic transcripts exhibited respectively higher or lower abundance in the fish fed with plant proteins (Tables 9 and 10).
Table 7 Hepatic transcripts exhibiting higher abundance in fish fed with vegetable oils after microarray analysis*

* Where P < 0·01 by t test (TIGR Multiple Experiment Viewer software; The Institute for Genomic Research, J. Craig Venter Institute, Rockville, MD, USA). The sixteen genes are classified following the P values.
Table 8 Hepatic transcripts exhibiting lower abundance in fish fed with vegetable oils after microarray analysis*

* Where P < 0·01 by t test (TIGR Multiple Experiment Viewer software; The Institute for Genomic Research, J. Craig Venter Institute, Rockville, MD, USA). The fifty-five genes are classified following the P values.
Table 9 Hepatic transcripts exhibiting higher abundance in fish fed with vegetable proteins after microarray analysis*

* Where P < 0·01 by t test (TIGR Multiple Experiment Viewer software; The Institute for Genomic Research, J. Craig Venter Institute, Rockville, MD, USA). The fifteen genes are classified following the P values.
Table 10 Hepatic transcripts exhibiting lower abundance in fish fed with vegetable proteins after microarray analysis*

* Where P < 0·01 by t test (TIGR Multiple Experiment Viewer software; The Institute for Genomic Research, J. Craig Venter Institute, Rockville, MD, USA). The sixty genes are classified following the P values.
Biological significance of the results: data clustering and gene ontology
We researched the significance of our data analysing some genes expressed in the clusters linked to specific biological process (Figs. 1 and 2). First, for the fish fed with or without fish oil, we observed three main clusters after analysing gene expression (Fig. 1 (a)). Cluster I was composed of numerous genes playing key roles in lysosomal and proteasomal proteolysis (cathepsins B and D, subunit of proteasomes) which were more expressed in the fish fed with vegetable oils (P < 0·01, t test; TMEV). We found also in this cluster one gene, CYP1A3, involved in xenobiotic metabolism. Cluster II was found to be mainly composed of genes involved in intermediary metabolism especially energy metabolism (succinate dehydrogenase, ubiquinol dehydrogenase) and lipid metabolism (fatty acid synthase (FAS) and long-chain fatty acid elongase). In this cluster, some genes also play roles in mRNA processing (splicing, maturation) (for details see Fig. 1 (b)). All the genes in cluster II were expressed at a lower level in the fish fed with vegetable oils (P < 0·01, t test; TMEV). Finally, cluster III was mainly composed of genes involved in cell growth and maintenance (keratin, kindlin, actin): they were expressed at a higher level in the fish fed with vegetable oils than in those fed the fish oil-based diet (P < 0·01, t test; TMEV). Overall, after dietary fish oil replacement by a blend of vegetable oils, a number of genes involved in lipid metabolism (lipogenesis, steroid synthesis, xenobiotic detoxification), protein catabolism, and transcription regulation were detected by gene ontology analysis (Table 11).

Fig. 1 (a) Hierarchical classification of differentially expressed genes between fish fed with or without fish oil (FO) (P < 0·01, t test, TIGR Multiple Experiment Viewer software; The Institute for Genomic Research, J. Craig Venter Institute, Rockville, MD, USA). Seventy-one hepatic genes were differentially expressed between the two dietary groups: sixteen were over-expressed (in red) and fifty-five were under-expressed (in green) in fish fed with vegetable oils (VO). (b) Detailed description of cluster II.

Fig. 2 (a) Hierarchical classification of differentially expressed genes between fish fed with or without fishmeal (FM) (P < 0·01, t test, TIGR Multiple Experiment Viewer software; The Institute for Genomic Research, J. Craig Venter Institute, Rockville, MD, USA). Seventy-five hepatic genes were differentially expressed between the two dietary groups: fifteen were over-expressed (in red) and sixty were under-expressed (in green) in fish fed with plant proteins (PP). (b) Detailed description of cluster IV.
Table 11 Major functional groups of hepatic transcripts exhibiting lower abundance in fish fed with vegetable oils after gene ontology analysis (GoMiner)*

* GoMiner software (Genomics and Bioinformatics Group, National Institutes of Health, Bethesda, MD, USA; http://discover.nci.nih.gov/gominer/).
Second, for the fish fed with plant proteins and fishmeal, four clusters were detected (Fig. 2 (a)). Whereas cluster I was composed of miscellaneous genes, cluster III was linked to intermediary metabolism, especially energy metabolism (quinone oxidoreductase, ubiquinol reductase), amino acid metabolism (arginase) and amino acid transport. Cluster II and cluster IV, even though also associated with metabolism, were focused on protein metabolism respectively with proteolytic (cathepsin, proteasome) and proteosynthetic (eight ribosomal proteins) pathways (for details, see Fig. 2 (b)). Clusters II, III and IV were composed of genes that were expressed much less in the fish fed with plant proteins (P < 0·01, t test; TMEV). Globally, the replacement of fishmeal by the plant protein sources indeed leads to a large number of genes involved in protein and amino acid metabolism being revealed by the gene ontology analysis (Table 12).
Table 12 Major functional groups of hepatic transcripts exhibiting lower abundance in fish fed with vegetable proteins after gene ontology analysis (GoMiner)*

* GoMiner software (Genomics and Bioinformatics Group, National Institutes of Health, Bethesda, MD, USA; http://discover.nci.nih.gov/gominer/).
Focus on specific differentially expressed genes
We focused the present study on specific genes that can illustrate the major pathways modified by the diet variation on rainbow trout liver. In the context of dietary fish oil replacement, five genes involved in proteolysis (cathepsin), energy metabolism in mitochondria (ubiquinol cytochrome c reductase), lipid metabolism (FAS and long-chain fatty acid CoA ligase) and detoxification metabolism (cytochromes P450) have been selected (Table 13). Using six individuals per dietary group and the quantitative RT-PCR, except for the cathepsin D, we confirmed that cathepsin B, ubiquinol cytochrome c reductase, FAS and cytochrome P4501A3 were expressed less in the fish fed with vegetable oils (P < 0·05; REST test). Moreover, we also checked that the cytochrome P4503A4 was more highly expressed in the liver of the fish fed with vegetable oils (P < 0·05; REST test). Also, in the context of dietary fishmeal replacement, genes involved in proteosynthesis (ribosomal proteins) and amino acid metabolism (glutamine synthetase) were selected to be analysed by quantitative RT-PCR (Table 14). We confirmed by quantitative RT-PCR that the expression of the glutamine synthetase gene was lower in the livers of the fish fed with plant proteins than in those fed fishmeal (P < 0·05; REST test): we analysed more precisely three glutamine synthase paralogous genes by specific amplification of the isoforms and we confirmed that GS01 and GS03 gene expressions were depressed by almost 7-fold in these fish. However, all the analysed genes coding for ribosomal proteins (four genes coding for ribosomal proteins of the 40S (6S and 7S) and 60S (L27 and L35) subunits) were not differentially expressed (P>0·05; REST test): these latter results did not confirm the microarray data.
Table 13 Selected genes analysed by real-time PCR: effect of dietary fish oil (FO) replacement by vegetable oils (VO)*

* Statistical differences in gene expression were evaluated between group means (six samples per group) by randomisation tests using the Relative Expression Software tool (REST©)(Reference Lopez, Rougemont, Loriod, Bourgeois, Loi, Bertucci, Hingamp, Houlgatte and Granjeaud23). The transcript level of target genes was normalised with EF1α-expressed transcripts.
† Positive and negative regulation means that the target gene is expressed at a higher or lower level, respectively.
Table 14 Selected genes analysed by real-time PCR: effect of dietary fishmeal (FM) replacement by plant proteins (PP)*
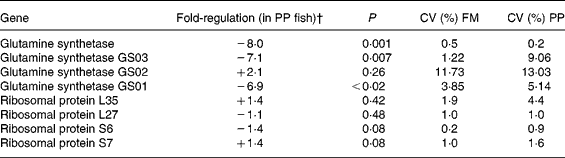
* Statistical differences in gene expression were evaluated between group means (six samples per group) by randomisation tests using the Relative Expression Software tool (REST©)(Reference Lopez, Rougemont, Loriod, Bourgeois, Loi, Bertucci, Hingamp, Houlgatte and Granjeaud23). The transcript level of target genes was normalised with EF1α-expressed transcripts.
† Positive and negative regulation means that the target gene is expressed at a higher or lower level, respectively.
Discussion
Overview
In the present study, the substitution of dietary fishmeal by vegetable proteins seems to have some adverse consequences on fish growth whereas no differences were found in fish fed vegetable oils as previously observed(Reference Gomez-Requeni, Mingarro, Calduch-Giner, Medale, Martin, Houlihan, Kaushik and Perez-Sanchez13, Reference Vilhelmsson, Martin, Medale, Kaushik and Houlihan14, Reference Richard, Kaushik, Larroquet, Panserat and Corraze18). This is the first ever set of nutrigenomics data in fish fed diets devoid of either fish oil or fishmeal. Analysis of the hepatic transcriptomes revealed that less than 100 genes were differentially expressed between all the nutritional conditions ( < 1 %), which is a relatively low number of differentially expressed genes. Two possible explanations are (i) that the cDNA microarray used in the present study is a generic rainbow trout cDNA tool (issue from 9023 cDNA extracted from different tissues at different developmental stages(Reference Govoroun, Le Gac and Guiguen21)), and not a specific rainbow trout liver cDNA microarray; and (ii) that although the diets differed in terms of ingredients (marine v. plant origins), they were not drastically different in terms of proximate composition (Tables 1–3). It is also interesting to note that when we analysed globally the data about differentially expressed genes, we observed no common genes between the two sets of experiments, with enrichment of differentially expressed genes in lipid metabolism and protein metabolism after fish oil and fishmeal replacements respectively. This suggests that the modification of liver transcriptomes was highly dependent of the origin of the ingredient, i.e. vegetable oil and plant proteins. Thus, in the following discussion, we will analyse separately the data from the two nutritional experiments by focusing on specific molecular actors.
Gene profiling after fish oil replacement by vegetable oil mixture: focus on specific genes
We found that replacement of fish oil by vegetable oils has a major impact on lipid, energy and xenobiotic metabolism.
As regards key actors involved in intermediary metabolism, we analysed FAS, which is the key enzyme of fatty acid biosynthesis in vivo; this metabolic pathway is highly active in rainbow trout liver(Reference Sargent, Tocher, Bell, Halver and Hardy32). Two FAS cDNA were spotted on the microarrays and both of them were detected (and clustered) to be down-regulated in the fish fed with vegetable oils and confirmed by quantitative RT-PCR. FAS gene expression was 10-fold lower in the fish fed with vegetable oils. The lower level of hepatic FAS gene expression in the trout fed with vegetable oils could be explained by the diets based on vegetable oils having higher levels of linoleic (18 : 2n-6) and linolenic (18 : 3n-3) acids than the fish oil(Reference Richard, Kaushik, Larroquet, Panserat and Corraze18) and by linolenic acid known to decrease FAS mRNA levels(Reference Jump, Clarke, Thelen and Liimatta33). The unambiguous finding in the present study of a significantly lower (10-fold) FAS gene expression in the fish fed with vegetable oils was not linked with a decrease of liver FAS activity in the same fish. This was even though FAS activity (IU/g liver) tended to be lower (P = 0·051) for the fish fed vegetable oils (4·0 (sd 0·9)) compared with fish fed fish oil (6·1 (sd 1·8)) (see Richard et al. (Reference Richard, Kaushik, Larroquet, Panserat and Corraze18)). It cannot also explain the modification of whole-body fatty acid composition which reflected mainly the composition of the diets (Tables 1 and 5). Our data about FAS mRNA levels suggest once more that molecular data (measures of gene expression level) are not always associated with significant effects at the protein-metabolic pathway level.
The second actor is one involved in lysosomal proteolysis, which includes proteases such as cathepsin B(Reference McGrath34). Cell proteins are always in a dynamic equilibrium between synthesis and degradation depending on nutritional status(Reference Mortimore, Kadowaki, Jefferson and Cherrington35). Lower expression of the CATB gene in the trout fed with vegetable oils cannot be presently explained and it is difficult to provide any putative biological consequence of this observation. The third gene is the ubiquinol cytochrome c reductase (UCR) which is involved in mitochondrial metabolism, i.e. oxidative phosphorylation. We found that the expression of UCR was down-regulated in the fish fed with vegetable oils. This is in agreement with the data of Barzanti et al. (Reference Barzanti, Battino, Baracca, Cavazzoni, Cocchi, Noble, Maranesi, Turchetto and Lenaz36) in rats, which showed a putative modification of UCR gene expression by dietary lipids.
Part of our analysis in the fish fed with vegetable oils dealt with the hepatic detoxification metabolism which is catalysed by a multi-enzyme family, namely the cytochromes P450s in rainbow trout(Reference Buhler and Wang-Buhler37). These enzymes have generally a large spectrum of endogenous as well as exogenous substrates, and CYP1A members catalyse the biotransformation of environmental disruptors or pollutants such as polychlorinated biphenyls in the liver before their elimination. Because CYP1A genes are induced by the presence of their substrates(Reference Buhler and Wang-Buhler37), the lower expression of the CYP1A3 gene in the liver of the trout fed diets with vegetable oils could be due to the lower levels of pollutants such as dioxins in these diets. This result is not surprising given that the fish oil is susceptible to contamination with lipophilic organic chemicals that are now ubiquitous in the marine ecosystems and consequently in aquaculture systems(Reference Jacobs, Covaci, Gheorghe and Schepens38, Reference Jacobs, Covaci and Schepens39). Our own results (G Corraze, unpublished results) show that the muscle levels of dioxins and polychlorinated biphenyls are reduced in trout fed vegetable oils compared with those fed fish oils (WHO toxic equivalent: 1·96 pg/g and 1·08 pg/g for fish oil and vegetable oil groups, respectively). The CYP3A27 in rainbow trout which metabolises testosterone can be reduced by phyto-oestrogen; however, this cannot presently easily explain the higher CYP3A27 gene expression in the fish fed vegetable oils. Moreover, CYP3A27 also has some similarities with the human CYP3A4(Reference Lee and Buhler40) which can convert cholesterol to 4-β-hydroxycholestrol before its elimination in bile salts(Reference Pikuleva41). Thus, the higher expression of the CYP3A27 gene in the rainbow trout fed with vegetable oils can be related to the lower level of plasma cholesterol observed in these fish (6·45 (sd 1·07) v. 3·97 (sd 0·23) g/l for fish fed fish oil and vegetable oils respectively; see Richard et al. (Reference Richard, Kaushik, Larroquet, Panserat and Corraze18)), suggesting that higher CYP3A27 activities can be the cause of the lower level of cholesterol in plasma. This needs further study to confirm and understand the link between fish oil replacement and the higher level of the CYP3A27 gene. Overall, the two hepatic cytochrome P450s described here can be proposed as molecular markers of dietary fish oil replacement by vegetable oils even though measures of enzymic activities will be necessary to confirm this status.
Gene profiling after fishmeal replacement by plant proteins: focus on specific genes
As the trout fed with plant proteins had significantly lower growth than the fish fed fishmeal, it was worth searching potential molecular actors to explain this phenotype, such as ribosomal proteins and glutamine synthetase.
A relatively high number of genes (n 8) coding for ribosomal proteins were detected in microarrays to be down-regulated in these fish. Moreover, we could classify them in the same cluster (IV) and considered that this set to be related to the lower growth of fish (for example, lower protein synthesis). Unfortunately, it was not possible to confirm these data by quantitative RT-PCR. We have no explanation to understand the discrepancy between the microarray and quantitative RT-PCR data. It is possible that these genes are false positive even though the data of microarrays seem unequivocal in humans (eight ribosomal genes with differential expression). An another explanation may be found in the specificity of the ribosomal genes: (i) there are more than seventy-nine ribosomal genes and, in trout, these are potentially in higher number due to pseudotetraploidy of the salmonids linked to a recent duplication of the genome(Reference Wool, Chan and Gluck42, Reference Meyer and Van de Peer43) and (ii) all these genes are not under the same control of their expression(Reference Ishii, Washio, Uechi, Yoshihama, Kenmochi and Tomita44). Thus, it is possible that we have not analysed the most appropriate ribosomal protein genes by quantitative RT-PCR. Overall, we prefer to take these data about ribosomal proteins with caution even though the growth rate and feed efficiency (which are major endpoints of the present nutritional experiments linked to the proteosynthesis potential) were unambiguously lower in the fish fed plant proteins than the fish fed fishmeal (P < 0·01).
We found that the glutamine synthetase mRNA levels were lower in the fish fed with plant proteins. Indeed, the lower glutamine synthetase gene expression (8-fold) observed in microarrays seems to be due to the lower levels of the two isoforms of the glutamine synthetase, i.e. GS01 (7-fold) and GS03 (6·9-fold), two genes highly correlated with rising levels of ammonia in rainbow trout(Reference Essex-Fraser, Steele, Bernier, Murray, Stevens and Wright45). Glutamine synthetase catalyses the transformation of glutamic acid into glutamine, leading to the elimination of ammonia(Reference Labow, Souba and Abcouwer46). Rainbow trout do have an active ammonia detoxification system, and glutamine synthetase activity increases after a meal naturally rich in proteins(Reference Wicks and Randall47) not only in the brain but also in the liver(Reference Essex-Fraser, Steele, Bernier, Murray, Stevens and Wright45). In the present study, the fishmeal replacement by plant proteins was associated with lower hepatic glutamine synthetase gene expression, thus potentially lowering the capacity of ammonia detoxification, possibly explaining the lower growth of these fish. However, the link between fishmeal replacement by plant proteins (naturally rich in glutamate) and lower glutamine synthetase gene expression is not clear and needs further study, especially at the level of enzyme activities.
Comparison with others nutrigenomic studies in fish
Very few studies have used nutrigenomics as a tool for the analysis of dietary fatty acids–gene interactions in aquaculture nutrition. Jordal et al. (Reference Jordal, Torstensen, Tsoi, Tocher, Lall and Douglas48) found regulation of several individual genes (for example, Δ6-desaturase, peroxysome proliferator-activated receptor, mitochondrial genes) after replacement of fish oil by 75 % rapeseed oil in Atlantic salmon. We do not find any common differentially expressed genes between the two studies. Indeed, we do not observe any change in Δ6-desaturase gene expression following the microarray analysis (data not shown) in contrast to Jordal et al. (Reference Jordal, Torstensen, Tsoi, Tocher, Lall and Douglas48), suggesting that the effects on gene profiling by vegetable oil replacement in fish may possibly vary with vegetable oil source and/or with fish species.
Concerning the analysis of dietary protein–gene interaction at the integrative level, an earlier study analysed the effect of dietary plant-protein substitution on hepatic metabolism using a proteomic approach(Reference Vilhelmsson, Martin, Medale, Kaushik and Houlihan14). The majority of the up-regulated proteins affected by the plant-protein diets were involved in energy production (NADPH, electron transferring flavoproteins). This is in contrast to data from the present study, in which lower expression levels of genes involved in energy metabolism were found. However, we should recognise that comparison between transcriptomic and proteomic data is difficult. A recent study by Salem et al. (Reference Salem, Silverstein, Rexroad and Yao49) on the hepatic gene expression profiles between fasted and fed rainbow trout showed an inhibition of protein synthesis gene expression (ribosomal protein) in fasted fish; these data can be related to ours on trout fed with plant-protein diets having reduced growth.
Conclusion
Our data based on a transcriptomic approach and confirmed by quantitative RT-PCR enable us to identify modifications of hepatic gene expression after intake of a plant-based diet by rainbow trout. This non-exhaustive list of genes could be useful and used in the future as powerful tools to more closely monitor the effects of the evolution of feeds used for farmed fish(Reference Naylor, Goldberg, Primavera, Kautsky, Beveridge, Clay, Folke, Libchenko, Mooney and Troell50).
Acknowledgements
We acknowledge M. J. Borthaire and C. Nunez for their excellent technical assistance. We thank F. Terrier, Y. Hontang and F. Sandres for rearing fish in the INRA experimental farm (Donzacq, Landes, France).
The present study was financed by the AGENAE French programme (Analyse du GENome des Animaux d'Elevage), CIPA (Comité Interprofessionnel des Produits de l'Aquaculture), OFIMER (Office National Interprofessionnel des Produits de la Mer et de l'Aquaculture), Aquitaine Region (no. CCRRDT-20051303004AB), 5th European projects (FAIR no. QLK5-2000-30068, ‘Perspectives of Plant Protein use in Aquaculture’ (PEPPA) and FAIR no. Q5RS-2000-30058, ‘Researching Alternatives to Fish Oils in Aquaculture’ (RAFOA)) and a 6th PCRD European project (contract no. 016249-2, ‘Sustainable Aquafeeds to Maximise the Health Benefits of Farmed Fish for Consumers’ (AQUAMAX)).
F. P. and D. E. were responsible for generating the trout microarrays. C. K. was co-responsible for generating statistical analysis of the microarray data. E. P.-J. generated the quantitative RT-PCR data. N. R., G. C., F. M. and S. K. conducted the nutritional experiments in fish. S. P. was responsible for project development, drafted the manuscript and is the corresponding author.
There are no conflicts of interest.


















