INTRODUCTION
Norovirus is a non-enveloped positive-sense, single-stranded RNA virus, which belongs to the family Caliciviridae. Since the first identification of Norwalk virus in 1972 [Reference Kapikian1], five norovirus groups and 32 genotypes have been found, and human strains cluster within GI, GII and GIV [Reference Patel2]. For norovirus, the faecal–oral route is the leading transmission mode. Contaminated water, food or other fomites are important transmission vehicles. Moreover, owing to its low infectious dose and large amounts of virus in faeces and vomitus, norovirus is highly contagious, and all age groups can be infected, especially younger children and the elderly. Norovirus infections can occur year round and outbreaks tend to peak during cold weather [Reference Glass, Parashar and Estes3].
Currently, norovirus is one of the most significant viral agents attributable to gastroenteritis. In sporadic gastroenteritis, 12% of mild and moderate cases in all age groups were norovirus infections [Reference Patel4]. In gastroenteritis outbreaks in aged-care facilities in Victoria, Australia, 65% were reported to be associated with norovirus, especially GII [Reference Bruggink, Dunbar and Marshall5]. Approximately 14% of norovirus-gastroenteritis outbreaks were estimated to be foodborne [Reference Verhoef6]. Waterborne norovirus outbreaks accounted for 1·5% [Reference Kroneman7], and have also been reported in different countries [Reference Altzibar8–Reference Riera-Montes10], including China [Reference Li11, Reference Zhou12]. Moreover, waterborne outbreaks were reported to be significantly associated with GI strains [Reference Matthews13].
Previous studies have reported some norovirus outbreaks in schools in eastern China [Reference Xue14, Reference Xu15]. In late 2014, a gastroenteritis outbreak occurred in a school in the city of Binzhou, China. Hundreds of students presented with the symptoms of diarrhoea and vomiting. After epidemiological investigation and pathogen detection, contaminated direct drinking water was identified as the transmission vehicle, and norovirus was strongly suspected as the pathogen causing this gastroenteritis outbreak. Here, we describe the investigation process and molecular characteristics of norovirus from this outbreak.
METHODS
Outbreak description and epidemiological investigation
On 25 November 2014, Shandong Center for Disease Control and Prevention (SDCDC) was informed that a gastroenteritis outbreak had occurred in a school in Binzhou. Hundreds of students developed diarrhoea and vomiting. Immediately, a control team including epidemiologists and microbial experts was formed to investigate this event. As a matter of course, the food supply was changed first on 25 November, and the drinking water supply was subsequently cut-off on 26 November. After these control measures, the number of patients decreased markedly (from 334 to 43 in 2 days). However, on 14 December, the school restored the drinking water supply. Days later, the gastroenteritis outbreak recurred. On 28 December the school again cut-off the drinking water supply and this outbreak eventually came to an end on 5 January 2015. A retrospective cohort study was performed to identify possible foodborne or waterborne risk factors on 26 November. The exposure was defined as consuming suspected foods or unboiled direct drinking water. A student with diarrhoea and/or vomiting (⩾3 times/day) was defined as a case. Non-cases were defined as students without symptoms of diarrhoea or vomiting at the longest incubation period of norovirus infection (48 h) after exposure to suspected risk factors. SPSS v. 20.0 (SPSS Inc., USA) was used to calculate the attack rate (AR), relative risk (RR), and 95% confidence interval (CI).
Environmental investigation
In order to recognize the possible contamination, an environmental survey on water source was conducted. Teachers were found to have been supplied with barrelled water, which was purchased and supplied by the school. Students were served direct drinking water. The construction log of water network and leaks in the water pipes, water heaters and surrounding facilities were also investigated. The network supplying direct drinking water was found to be buried at a depth of 0·7 m and had been used for 2 years. Moreover, several septic tanks were close to the network (only 1 m away), and the distance between direct drinking-water supply network and sewage pipes was <2 m.
Samples collection
At the peaks of two outbreak waves (25, 26 November and 21, 22 December), a total of 33 clinical specimens (including six vomitus and 27 stool specimens) were collected from confirmed cases by health professionals. At that time, every food item was aseptically collected from canteens daily and 46 food samples were collected eventually. At the second outbreak wave (22 December), five direct drinking-water samples were aseptically collected from different sites in the direct drinking-water supply network. Clinical specimens were frozen, and food and water samples were stored at a cold temperature of 4 °C.
Detection of pathogenic bacteria
Common foodborne pathogenic bacteria from clinical specimens, food and drinking water samples were examined. The detection and confirmation of Staphylococcus aureus, Bacillus cereus and Salmonella spp. in all samples was according to the AOAC official method (995·12, 980·31, 978·24), and diarrhoeagenic Escherichia coli was isolated and confirmed according to the reference [Reference Ben Salem-Ben Nejma16]. Furthermore, in drinking water samples, microbiological indexes for drinking water quality (aerobic plate count, coliforms, Pseudomonas aeruginosa) were detected according to the AOAC official method (990·12, 998·08) and ISO standard (16266:2006).
Detection of gastroenteritis viruses
Sample processing
For each clinical specimen, phosphate-buffered saline was added to obtain a 10% (w/v) suspension, which was centrifuged at 4000 g for 30 min at 4 °C. The supernatant fluid was used to extract viral nucleic acid.
For each water sample (800 ml), MgCl2 was added to a final concentration of 0·05 m, and pH was adjusted to 3·5 by HCl. Then, water samples were filtered through a mixed cellulose ester membrane filter (pore size 0·45 µm, Advantec, Japan). Ten millilitres of 3% beef extract solution (pH 9·0) was used to elute viruses from the membranes by ultrasonication for 5 min.
Nucleic acid extraction and polymerase chain reaction (PCR) of gastroenteritis viruses
To detect gastroenteritis viruses (rotavirus group A, norovirus GI and GII, astrovirus, sapovirus, adenovirus), the supernatant fluid of clinical specimens and concentrated water samples were used to extract viral RNA and DNA using the QIAamp viral RNA mini kit and QIAamp DNA mini kit (Qiagen, USA), respectively, according to the manufacturer's protocols. Reverse transcription–PCR (RT–PCR) was performed using the Access RT–PCR System (Promega, USA) to amplify rotavirus group A, norovirus GI and GII, astrovirus and sapovirus sequences. PCR were performed to amplify adenovirus sequences using Roche Taq DNA polymerase (Roche, USA). Primers were according to previous studies [Reference Gómara17–Reference Xu, McDonough and Erdman22].
TA cloning
The positive RT–PCR products from water samples were gel-purified by QIAquick gel extraction kit (Qiagen) and ligated into the TA cloning vector (pGEM-T Easy vector; Promega). The ligation products were transformed into competent Escherichia coli JM109 cells by the heat-shock method. After blue and white screening, ten positive clones were selected.
Sequencing, homologous comparison and phylogenetic analysis
All positive PCR products of clinical specimens and positive clones from water samples were sequenced by the BigDye Terminator v. 3.1 Cycle Sequencing kit (Applied Biosystems, USA), on an ABI 3130 genetic analyser (Applied Biosystems). The amplified sequences of norovirus GI and GII correspond to nucleotide position 5342–5671 of strain Norwalk/68/US (accession no. M87661) and nucleotide position 5003–5389 of strain Lordsdale/93/UK (accession no., X86557), respectively. Genotyping was performed by the online Norovirus Genotyping Tool v. 1.0 [Reference Kroneman23]. Homologous comparison and phylogenetic analysis were performed among norovirus sequences from this study and reference strains downloaded from GenBank. Nucleotide sequence alignments were performed by BioEdit v. 7.0.0 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). MEGA v. 5.0 (http://www.megasoftware.net/) was used to construct the phylogenetic tree by the neighbour-joining method. Bootstrapping test was performed with 1000 replications.
Nucleotide sequence accession numbers
Nucleotide sequences obtained from this outbreak were deposited in the GenBank database with accession numbers KR611787–KR611793 and KR706448–KR706461.
RESULTS
Epidemiological investigation
The first case was recognized on 17 November 2014. Up to 5 January 2015, a total of 1614 cases were identified. Figure 1 shows the distribution of cases by date of symptom onset. Of 926 patients with complete case information, 56·5% (523/926) were males. Only 0·76% (7/926) were teachers. Of 406 cases diagnosed at the school clinic, nausea (80·1%) was reported to be the most common symptom, followed by abdominal pain (69·7%), vomiting (66·5%), diarrhoea (56·4%) and headache (52·2%). Symptoms in most cases were mild, and cases recovered 1–2 days after treatment. In total, 402 individuals were included in the retrospective cohort study. No significant difference of AR was observed in 12 foodborne risk factors. However, the AR was significantly higher in those exposed to unboiled direct drinking water than those unexposed (38·5% vs. 28·1%). Table 1 shows the results of statistical analysis for suspected risk factors.
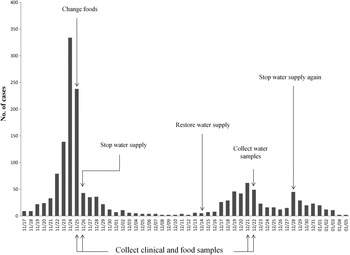
Fig. 1. Distribution of cases by date of symptom onset.
Table 1. Results of statistical analysis for suspected risk factors

AR, Attack rate; RR, relative risk; CI, confidence interval.
Detection results of gastroenteritis viruses
Rotavirus group A, astrovirus, sapovirus and adenovirus were not detected in clinical specimens and drinking water samples. Nevertheless, norovirus was positive in 12 clinical specimens (36·3%, 12/33). GII.4 was the most predominant genotype (50%, 6/12), followed by GI.2 (41·7%, 5/12) and GII.6 (8·3%, 1/12). Furthermore, in one water sample collected from a dormitory building, norovirus was detected. After TA cloning, ten positive clones were selected and sequenced. Finally, nine norovirus sequences and one bacterial sequence from non-specific amplification were obtained. These nine norovirus sequences belonged to four different genotypes, including GII.13 (66·7%, 6/9), GII.4 (11·1%, 1/9), GI.1 (11·1%, 1/9) and GI.6 (11·1%, 1/9). Table 2 shows the results of sample collection and detection.
Table 2. Results of samples collection and detection

* Detected bacterial pathogens include Staphylococcus aureus, Bacillus cereus, Salmonella spp. and diarrhoeagenic Escherichia coli.
† NT, samples were not tested.
Homologous comparison and phylogenetic analysis
Figure 2 shows the phylogenetic relationship of norovirus sequences from this study and reference strains. Five GI.2 sequences from this study displayed 99·3–100% nucleotide identity to each other, and 98·6–99·3% identity to another Chinese reference strain (KF306212). GI.6 sequence displayed 99·3% and 100% nucleotide identity to a Chinese strain (KM249616) and a Japanese strain (AB901031), respectively. GI.1 sequence showed 99·3% and 99·6% nucleotide identity to a Chinese strain (HM241691) and a Japanese strain (AB522413), respectively. Six GII.4 sequences from clinical specimens had 96·7–100% nucleotide identity among themselves, and displayed 97·0–100% nucleotide identity to a GII.4 sequence from a water sample. Additionally, all GII.4 sequences from this study were closely related to GII.4 2012 variant (JX459908), with 97·6–99·4% nucleotide identity, and displayed 97·3–99·1% nucleotide identity to a GII.4 2012 variant from China (KF306214). GII.13 sequences displayed 99·0–100% nucleotide identity to each other, and showed a close relationship (98·5–98·8% identity) with strains isolated in Germany (KC832470). GII.6 sequence had 95·5% identity with a Chinese reference strain (JX989075), and 97·3% identity with a Japanese reference strain (AB504694).
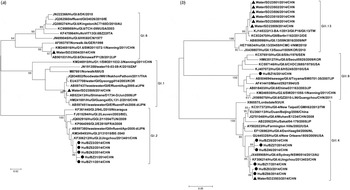
Fig. 2. Phylogenetic relationships of norovirus GI (a) and GII (b) sequences from this outbreak and reference strains. ● and ▲ indicate norovirus sequences obtained from cases and water sample, respectively. The phylogenetic trees were constructed based on 291-nt GI sequences (position 5362–5652 on strain Norwalk/68/US) and 338-nt GII sequences (position 5029–5366 on strain Lordsdale/93/UK), respectively. Reference strains are as follows: GI: JQ934802, JN191358, KF361440, FJ515294, KF306212, KC998959, JQ388274, KM246910, KM349493, JQ692916, KP064095, JN699045, JN222366, KF475964, M87661, KM246916, AB597380, AB901031, JQ362560, AF093797, HM241691, AB597451, EU437708, AB597437, AB522413. GII: HE716634, JX439807, KC517372, KM246935, KC597146, AB818403, KC576910, HM635127, AY502023, AB220922, EF126962, EU366113, GU445325, JX459908, AB809998, KJ407072, X86557, JX989075, AB504694, AF414410, JQ751048, KF306214, KJ145322, KC832470.
DISCUSSION
This study describes an investigation process of a gastroenteritis outbreak in China. Based on the epidemiological, environmental and microbiological investigations, direct drinking water contaminated by norovirus was responsible for this outbreak. This conclusion was supported by several evidences.
First, symptoms of cases (a short duration of diarrhoea and vomiting) were in accord with the clinical features of norovirus infection [Reference Robilotti, Deresinski and Pinsky24]. After bacteria detection, all clinical samples were negative for common foodborne pathogenic bacteria. Moreover, after detection of gastroenteritis viruses, only norovirus was detected. Further, this outbreak occurred in winter, following the seasonal pattern of norovirus outbreak. The above-mentioned facts revealed that this gastroenteritis outbreak was attributed to norovirus. Second, no foodborne risk factor was identified. Nevertheless, the AR was significantly 1·37 times higher in individuals exposed to unboiled direct drinking water than those unexposed. Additionally, teachers, who received a different water source (barrelled water) than the students, made up an extremely small proportion (0·76%) in all cases. All of this indicated that unboiled direct drinking water was the risk factor for this outbreak. Third, four water samples from dormitory and teaching buildings were up to national standards for drinking water quality. However, in one water sample collected from the machine room, the aerobic plate count, coliforms exceeded the upper limit (2·7 × 107 c.f.u./ml and 1·4 × 105 c.f.u./ml). P. aeruginosa was also positive in this sample, which should not be detected. Moreover, norovirus was positive in one water sample from the dormitory building, and GII.4 sequences from this water sample and clinical specimens showed a close relationship. We also found that the direct drinking-water supply network was close to several septic tanks and sewage pipes. These results indicated that the drinking water was contaminated and acted as the transmission vehicle for norovirus infection.
After being informed of this outbreak, subsequent controls measures were taken. The food and water supply were changed. Drinking water for students was supplied by standby well. Cases were isolated and treated, and strict disinfection of excreta and medical waste from cases was performed to avoid cross-infection. Hand hygiene and environmental disinfection practices were implemented. Health education was also carried out to improve students' knowledge of norovirus infection.
However, in spite of the suspicion of drinking water, following the epidemiological investigation there was no direct evidence indicating that the water was contaminated. Therefore, disinfection of direct drinking water was not conducted in a timely manner. Moreover, stored well water was not sufficient to supply students for long, and the school had to restore the suspected direct drinking water, which caused the occurrence of the second outbreak wave. The drinking water supply was turned off again until it was confirmed to be contaminated by the detection of norovirus, and this outbreak eventually came to its conclusion. Subsequent disinfection of drinking water was strictly implemented.
This outbreak revealed some issues on management and control of a norovirus outbreak. First, this case was not reported in time, meaning control measures were not taken immediately. For example, the water supply was not turned off on the day of outbreak. Therefore, the reporting system should be improved. Second, this outbreak suggested that monitoring systems on drinking water were imperfect, and disinfection of drinking water was not conducted in time after the first outbreak wave. In future, monitoring systems should be strengthened to prevent future occurrences, and disinfection of drinking water should be strengthened and normalized. Third, strengthening of stored drinking water is essential in future.
Interestingly, co-circulation of multiple norovirus genotypes was observed in this case, revealing that recombination may occur. RNA recombination is common in norovirus, and norovirus types tend to co-circulate before the emergence of new types. It has been demonstrated that almost all contemporary non-GII.4 noroviruses are recombinant viruses and recombination can allow norovirus to escape from herd immunity [Reference White25]. Therefore, co-circulation and potential recombination of norovirus may accelerate this gastroenteritis outbreak. For public health, surveillance on co-circulation of norovirus types should be strengthened.
All GII.4 sequences in this case belonged to GII.4 variant Sydney 2012, proving that GII.4 variant Sydney 2012 circulated in Shandong Province. Previous reports have demonstrated that waterborne norovirus outbreaks were more likely to be caused by GI [Reference Matthews13, Reference Lysén26]. Thus, GI genotypes may have played an important role in this case. GII.13 was the most frequent detected genotype in water samples in this study. A previous study reported that 6·4% of norovirus outbreaks in aged-care facilities were associated with GII.13, second to GII.4 variants [Reference Bruggink, Dunbar and Marshall5]. Therefore, GII.13 may also play a significant part and remains to be investigated.
Vomiting is an obvious symptom in cases of this outbreak, and infectious aerosols can be generated from vomitus. Nevertheless, it is difficult to distinguish primary and secondary cases, which reveals little about person-to-person transmission in this outbreak. This is a limitation of this study. Furthermore, norovirus was not tested for in food samples, and the difficulty of norovirus detection in water samples meant that some norovirus genotypes in clinical and water specimens were unaccounted for in this outbreak.
In conclusion, this study describes a gastroenteritis outbreak associated with a direct drinking-water system in China, which improves our knowledge on waterborne norovirus outbreaks, and provides many prevention and control practices for policy-makers. Current measures should be changed to better manage gastroenteritis outbreaks. In the future, strengthening water supply management is vital, and disinfection measures should be normalized. More important is that, as reported in previous study [Reference Xue14], relevant departments should enhance publicity and education to improve the public's awareness of norovirus gastroenteritis prevention.
ACKNOWLEDGEMENTS
We acknowledge the assistance from the staff of SDCDC and Binzhou CDC. This study was supported by a grant from the National Natural Science Foundation of China (81 573 209) and a grant from the Shandong Provincial Natural Science Foundation, China (ZR2014HM076).
DECLARATION OF INTEREST
None.







