INTRODUCTION
Suid herpesvirus 1 (SuHV1), or pseudorabies virus (PrV), a member of the family Herpesviridae, subfamily Alphaherpesvirinae, genus Varicellovirus, is the causative agent of Aujeszky's disease [Reference Pomeranz, Reynolds and Hengartner1], a notifiable disease in swine of great economic importance. In spite of its broad host range which includes nearly all mammals except higher primates and humans, members of the Suidae family (domestic and feral pigs, wild boar) are the only hosts able to survive a productive infection and serve as a virus reservoir [Reference Kluge, Straw, D'Allaire, Mengeling and Taylor2]. In domestic pigs the use of gE-deleted marker vaccines in combination with DIVA (differentiating infected from vaccinated animals) strategies for serological testing, and strict control efforts has led to substantial improvement of the disease situation and eventually in the elimination of PrV from many parts of Europe and North America [Reference Mettenleiter3].
However, it soon transpired that despite the tremendous success of the vaccination schemes in domestic pig populations, the complete eradication of PrV could not be achieved. Early reports from the mid-1980s and the beginning of the 1990s have already provided evidence for PrV infections in free-roaming feral pig and wild boar populations in The Americas and also in Europe and even North Africa. In Europe, the detection of seropositive wild boar in Germany, Italy, the former Yugoslavia and The Netherlands, indicated that PrV had found an ecological niche already in times when national eradication programmes were established in many countries (for review see [Reference Müller, Conraths and Hahn4]). In recent years, increased efforts in serological surveys and monitoring indicated the occurrence of PrV infections in a number of other European countries, e.g. Croatia, the Czech Republic, France, Slovenia, Poland, Russia, Switzerland and Spain with seroprevalences ranging between 2·4% and 56·25% [Reference Szweda5–Reference Sedlak, Bartova and Machova12]. These studies showed that PrV infections were more widely distributed in wild boar in Europe than previously assumed.
Isolation and characterization of the virus from infected wild boar should help to identify the origin of infections. Most PrV of wild boar origin were supposedly obtained from dead hunting dogs or wild carnivores which had contact with PrV-infected wild swine and subsequently succumbed to infection [Reference Glass13]. This view is supported by reports describing the detection of PrV in brain tissue of a Florida panther (Felis concolor coryi) and in hunting dogs that suffered from clinical symptoms typical for Aujeszky's disease in the USA, Austria and Belgium, respectively [Reference Glass13–Reference Cay and Letellier15].
However, only few attempts were made to detect, isolate and characterize viruses from the wild reservoir host in regions where PrV is endemic in feral pigs or wild boar. In the USA, PrV isolates of feral pig origin were used for experimental studies [Reference Hahn16–Reference Romero19]. In Europe, PrV isolated from wild boar in Italy, Germany and Belgium has been analysed [Reference Cay and Letellier15, Reference Capua20–Reference Lutz23]. Usually, molecular characterization of PrV is performed by analysis of restriction fragment length polymorphisms (RFLP) using BamHI delineating four major genome types [Reference Herrmann, Heppner and Ludwig24]. Using this technique, the few available PrV isolates of wild boar origin which were obtained from endemic regions in the USA, Italy and Germany showed typical features of a PrV type I genome [Reference Müller22, Reference Lutz23, Reference Hahn, Peirce and Gibbs25, Reference Capua26]. Moreover, PrV DNA was detected using polymerase chain reaction (PCR) in organ samples from free-roaming wild boar in Spain [Reference Ruiz-Fons27]. Considering the large spread of PrV infections in wild boar populations across Europe and the changing disease situation in domestic swine, there is an urgent need to increase our understanding of the epidemiology of the infection in wildlife and to provide tools to trace possible spillover into domestic pigs or other livestock. Thus, it is still unknown how many PrV variants are circulating among wild boar across Europe and how they are related to each other. In order to address these questions we set out in a multi-national approach to isolate and characterize PrV isolates of wild boar origin from different parts of Europe.
MATERIALS AND METHODS
Collection of PrV isolates and DNA
Between 1993 and 2008, PrV isolates of wild boar origin or DNA of PCR-positive wild boar were collected for molecular characterization from Germany, France, Slovakia, Hungary and Italy either during targeted virological surveys on PrV in regions where PrV infections were shown to be endemic in free-ranging wild boar, or during routine virological screening on the presence of classical swine fever virus (CSFV). Additionally, PrV isolated from hunting dogs that suffered from Aujeszky's disease probably contracted by direct contact with wild boar in endemic regions were included in the study. Depending on the design of the virological survey conducted in different countries and the methods established in the respective laboratories, PrV was isolated from organ tissues, e.g. lung, tonsils, spleen, kidney, lymph nodes (wild boar), brain (hunting dogs), or from nasal and genital swabs (wild boar) as previously described [Reference Romero19, Reference Müller22, Reference Lutz23]; using African Green Monkey (Vero), PK-15, SPEV or SK6 cells. Cell cultures were screened for a cytopathic effect (CPE) indicative of herpesvirus infection and PrV antigen was detected by indirect peroxidase immunostaining using a goat polyclonal anti-PrV antibody and AEC as substrate. Cell cultures were passaged at least four times to yield sufficient amount of virus for subsequent characterization.
From Spain, only DNA from PCR-positive wild boar was available as there was no laboratory capacity to isolate PrV. The DNA was extracted using commercial DNA extraction kits (Machery & Nagel, Germany) from trigeminal ganglia and tonsils of wild boar from regions in central Spain where PrV infections are endemic in wild boar [Reference Ruiz-Fons27].
Where available, the species that the virus was isolated from, the location (municipality/region), and year of isolation were recorded. All PrV isolates or DNAs were submitted for further molecular characterization and analysis to the National Reference Laboratory for Aujeszky's disease at the Friedrich-Loeffler-Institut, Germany.
Virus identification
Virus identification was performed by direct fluorescence antibody test (FAT) using a polyclonal FITC-labelled anti-PrV serum (AV-Gammacon, Czech Republic [Reference Müller22]) or by conventional PCR using primers specific for the gene encoding the glycoprotein D (gD) of PrV with slight modification to the method previously described [Reference Schang and Osorio28]. Briefly, after DNA extraction using a commercial kit (Machery & Nagel) we used a LightCycler-adapted temperature profile, i.e. amplification of the 217-bp fragment of the gD gene was performed for 40 cycles (15 s at 95°C, 10 s at 60°C, 10 s at 72°C).
RFLP
Conventional typing of PrV isolates was conducted by RFLP using BamHI [Reference Müller22]. Briefly, PrV isolates were propagated in Vero cells for at least 2 days to reach a sufficient virus titre. Subsequently, DNA was extracted from cells and supernatants with PMQN [phenol, m-cresol, 8-hydroxychinoline, 0·15 m NaCl (pH 8·0)]/chloroform/isoamylalcohol (50/48/2), resuspended in TE buffer [10 mm Tris–HCl, 1 mm EDTA (pH 8·0)] and stored at −20°C. For restriction enzyme analysis, viral DNA was digested with BamHI (Life Technologies, Germany) at 37°C for 1·5–3 h. DNA fragments were electrophoretically separated in 0·7% agarose gels (Life Technologies) at 50 V overnight and stained with ethidium bromide. Classification of DNA fragment patterns was performed using the PrV strains Kaplan and Bartha as reference as previously described [Reference Müller22, Reference Herrmann, Heppner and Ludwig24]. DNA from Spanish wild boar had to be excluded from RFLP analysis because of the limited amount of DNA available from organs.
Sequencing
Sequencing was performed after amplification of an ∼800-bp fragment comprising a part of the gC open reading frame (start codon at nt 54029) and the 5′ non-coding region by PCR with primers gC-For (5′-GTGCGCCACTAGCATTAAATCCGT-3′, nt 53933-53956; Genbank accession no. BK00174) and gC-Rev (5′-CTGTACAGGAGGAGCGAGACGTT-3′, nt 54732-54710) using pfx DNA polymerase (Invitrogen, Germany) in a volume of 25 μl with the provided reagents. Amplification reactions were performed in a thermocycler (MWG, Germany) under the following conditions: after an initial incubation step for 3 min at 95°C and 2 min at 72°C, 35 cycles of 30 s at 95°C and 2 min at 72°C were performed with a final elongation step for 4 min at 72°C and an indefinite incubation at 4°C. PCR products were separated on 1% agarose gels, purified and sequenced on both strands using the Big Dye R terminator cycle sequencing kit (Applied Biosystems, Germany) with the primers used for amplification. Sequence determination was performed with the genetic analyser 3130 (ABI).
Alignment and phylogenetic analysis
Sequences encompassing 732 nt coding for 223 amino acids in total were aligned and their evolutionary history was inferred using the neighbour-joining method [Reference Saitou and Nei29] as implemented in MEGA4 [Reference Tamura30]. For the deduced amino-acid sequences, the evolutionary distances were computed using the Dayhoff matrix-based method [Reference Schwarz, Dayhoff and Dayhoff31]. All positions containing gaps and missing data were eliminated from the dataset (complete deletion option). The bootstrap test as percentage of replicate trees (500 replicates) in which the associated taxa clustered together was used as previously described [Reference Felsenstein32]. Nucleotide sequences generated in this study were submitted to GenBank. Accession numbers are given in Table 1.
Table 1. PrV isolates of wild boar origin included in the study
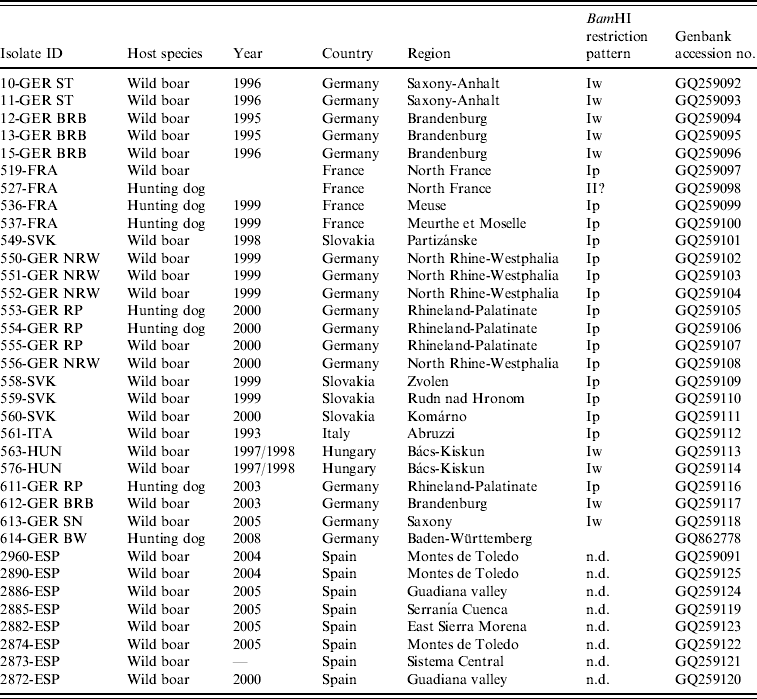
n.d., Not determined.
RESULTS
Between 1993 and 2008, a total of 27 PrV isolates of wild boar origin were obtained, of which 20 were isolated directly from PrV-infected wild boar and seven from hunting dogs. Sixteen isolates originated from southwestern or northeastern Germany.
Four PrV isolates were obtained from northwestern regions in France or central or southern regions of Slovakia, respectively. PrV from Hungary (n=2) were isolated in regions adjacent to the border to Serbia. The wild boar PrV isolate from Italy was identical with one previously described [Reference Capua26]. From Spain, eight DNA samples from seven different districts where PrV infections are known to occur were selected and included in the phylogenetic analysis (Table 1, Fig. 1).

Fig. 1. Origin of pseudorabies virus (•) isolated from wild boar and hunting dogs.
RFLP
The BamHI-induced RFLP of genomic DNA of PrV isolates from wild boar showed that all but one belonged to PrV genotype I. The BamHI restriction pattern of isolate 527-FRA differed considerably from those of the reference strains Bartha and Kaplan, and showed typical features of PrV genotype II, in particular the absence of BamHI fragment 2 (Table 1). Two different subtypes were evident within PrV genotype I. Most of PrV isolates (n=17) showed BamHI restriction patterns similar to the standard pattern as exhibited by PrV strain Kaplan identifying them as subtype Ip. In contrast, the BamHI restriction patterns of four wild boar isolates from Brandenburg (612-GER BRB) and Saxony (613-GER SN), Germany, and from the Bács-Kiskun district in Hungary (563-HUN, 576-HUN) exhibited the same pattern as wild boar PrV previously isolated in eastern Germany (10-GER ST, 11-GER ST, 12-GER BRB, 13-GER BRB, 15-GER BRB) which had been characterized as subtype Iw [Reference Müller22]. Due to the loss of a BamHI site in the inverted repeat regions, they are also referred to as 5-10/5-12 double fusion strains (Fig. 2). With the exception of the absence of fragment 2, 527-FRA showed a similar BamHI restriction pattern as the isolates from eastern Germany and Hungary.

Fig. 2. BamHI restriction pattern of viral pseudorabies virus (PrV) DNA of selected wild boar PrV. kb, kilobase DNA ladder; lane 1, isolate 553-GER RP; lane 2, isolate 554-GER RP; lane 3, isolate 550-GER RP; lane 4, isolate 612-GER BRB; lane 5, isolate 563-HUN; lane 6, isolate 527-FRA; lane 7, isolate 519-FRA; lane 8, isolate 614-GER BW; lane 9, isolate 549-SVK; lane 10, isolate 559-SVK; lane 11, PrV reference strain Bartha; lane 12, PrV reference strain Kaplan. Horizontal arrows indicate 5/10-5/12 fusion bands and loss of DNA fragments 10 and 12 (lanes 4–6). Vertical arrows show loss of DNA fragment 2 (lane 6) and fragment 7 (lane 11). The latter is characteristic for the PrV reference Bartha.
Sequence and phylogenetic analysis
Sequence analysis revealed about 40 variable positions within the 732-bp fragment spanning 61 nt in the 5′ non-coding region of the gC gene and part of the open reading frame encoding the N-terminal 223 amino acids of gC. Phylogenetic comparison of the nucleotide and the deduced amino-acid sequences revealed almost identical results and identified at least eight different PrV variants circulating in wild boar populations in Europe indicating a separation into two distinct clades, designated as ‘A’ and ‘B’ (Fig. 3).
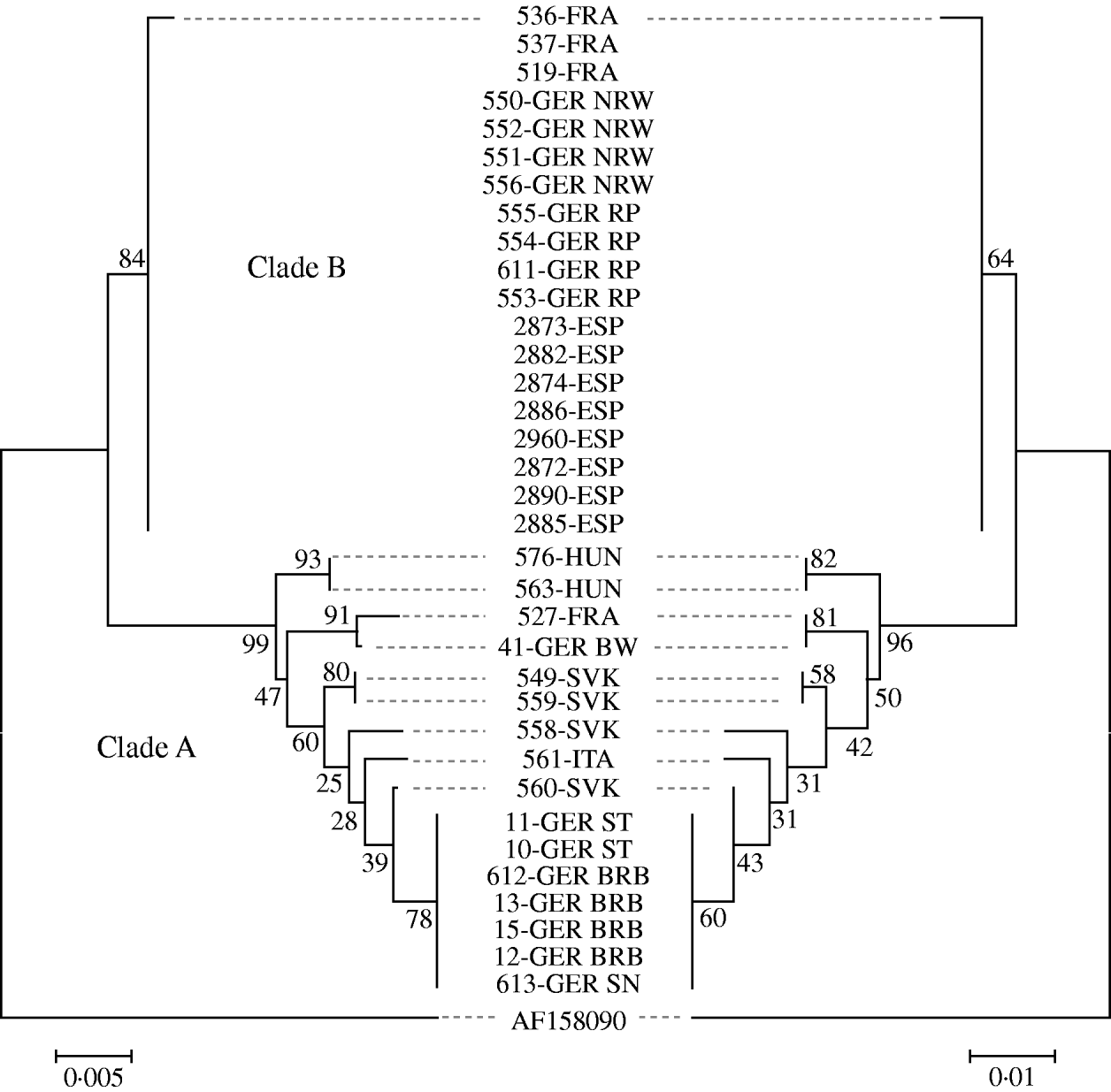
Fig. 3. Phylogenetic trees of the evolutionary history inferred from the glycoprotein C gene sequences (left) and the deduced amino-acid sequences (right) using the neighbour-joining method. Bootstrap values (500 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances. The Chinese strain Ea (AF 158090) was used as outgroup.
Clade B comprised 19 isolates from Germany (North-Rhine Westphalia, Rhineland-Palatinate), France and Spain. With the exception of isolate 536-FRA which displayed a single amino-acid substitution, all sequences within this group were identical. All other isolates formed an apparently more variable clade A comprising isolates from Germany (Baden-Württemberg, Brandenburg, Saxony, Saxony-Anhalt), Slovakia, Hungary, Italy and France. Within clade A, sequences of isolates from the northeastern parts of Germany formed one homogenous group, while for the remaining sequences no further clusters were obvious. Based on both nucleotide and amino-acid sequences, the PrV isolates from Slovakia fell into three different groups. The sequences of the two isolates from Hungary were identical. Isolate 527-FRA seems closely related to isolate 614-GER from Baden-Württemberg, Germany (Fig. 3).
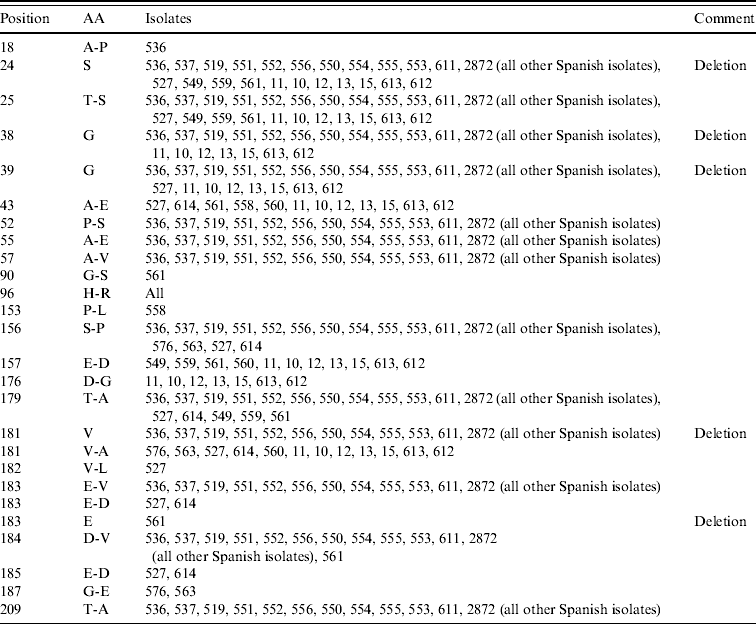
An alignment of the deduced amino-acid sequences showed insertions/deletions (indel) at positions 18, 38–39, 181 and 183 (Table 2). The indel at position 181 is unique to clade B. Further, 561-ITA was the only isolate were an indel was found at position 183 (Fig. 4).
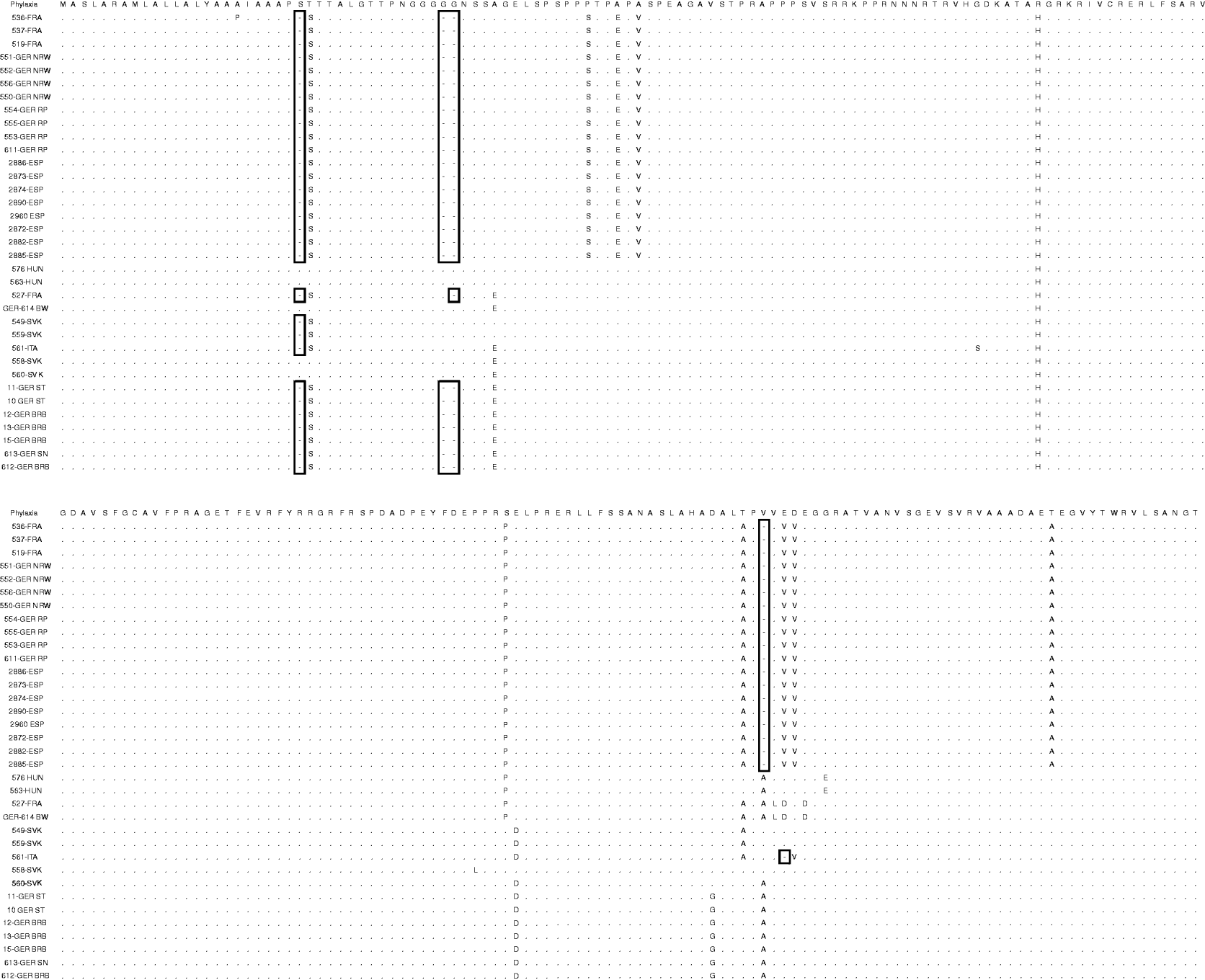
Fig. 4. Alignment of the deduced amino-acid sequences of gC in comparison with the Phylaxia pseudorabies virus strain. Indels are indicated (open boxes).
Table 2. Summary of variable amino acids (AA) in gC in comparison with the reference strain Phylaxia
DISCUSSION
In this study, a large set of European PrV isolates of wild boar origin was analysed. It comprised 35 isolates from wild boar or hunting dogs from Germany, France, Spain, Italy, Slovakia and Hungary isolated between 1993 and 2008. Prior to this study, PrV isolations from wild boar and subsequent virus characterization had only been reported from Italy, France and Germany [Reference Capua20–Reference Lutz23]. Our findings not only confirm the presence of PrV in wild boar populations of France, Germany, Spain and Italy but also suggest a far more widespread circulation of PrV, even in countries which have not yet reported any evidence for PrV infection in wild boar, e.g. Slovakia and Hungary. Since several of the investigated isolates were obtained from border areas, e.g. between Germany/Poland, Germany/Belgium, Germany/Czech Republic, Hungary/Serbia, and Hungary/Croatia, the detection of PrV in these areas could be the result of migrating wild boar. It is also likely that PrV might be also prevalent in additional countries in wildlife. However, this needs to be confirmed by serological studies.
In our study, we compared conventional RFLP and partial sequencing in the analysis of PrV isolates of wild boar origin to gain more insight into the epidemiology of PrV infections in wild boar in Europe. The results obtained by either technique did not correlate well as has been observed by others (see e.g. [Reference Goldberg33]). Conventional RFLP of genomic DNA using BamHI has initially been applied as the standard method for the characterization of PrV and for inferring genetic relationships in them [Reference Herrmann, Heppner and Ludwig24]. We used this method for comparison with results obtained in previous studies. It resulted in a first classification of the PrV isolates of wild boar origin and showed that all isolates, except one, belonged to genotype I (Fig. 2). This confirms results obtained with PrV isolates of wild boar or feral swine from Italy, Germany and North America [Reference Capua20, Reference Müller22, Reference Hahn, Peirce and Gibbs25]. Interestingly, wild boar isolates from Germany and Hungary showed the same rare PrV subtype Iw (5-10/5-12 double fusion strains) confirming previous associations with PrV isolates from domestic pig and cats in Hungary [Reference Nagy34–Reference Müller36]. Since genotype II strains prevailed in domestic pigs in central Europe in recent years, it has been speculated that PrV infections of wild boar must have occurred some time ago [Reference Capua20]. This may indicate that the sole genotype II wild boar isolate 527-FRA had been introduced into wild boar from domestic pigs only recently.
RFLP is inexpensive and can detect variation across large genomic regions [Reference Herrmann, Heppner and Ludwig24]. However, it detects only a small proportion of the total genetic variation. Therefore (partial) DNA sequencing has been introduced in the investigation of Aujeszky's disease outbreaks in domestic pigs in the USA [Reference Goldberg33]. However, while sequencing yields detailed information about the genetic variation in the targeted genomic region, it lacks information on the genome as a whole [Reference Goldberg33]. We sequenced part of the 5′ non-coding and coding regions of the gene encoding gC. This protein is a non-essential component of the virion envelope and is involved in the attachment of the virion to the cell [Reference Mettenleiter3]. It is considered as a more variable part of the PrV genome due to genetic alterations as a result of antibody selection [Reference Ishkawa37]. Partial sequence analysis of the gC gene region also allowed a clear differentiation between isolates exhibiting an identical BamHI restriction pattern (Fig. 2) [Reference Ishkawa37]. At least eight different PrV sequence types were shown to occur in wild boar in Europe with up to three circulating in countries such as Germany and Slovakia. Interestingly, the wild boar PrV isolates from southwestern Germany, the eastern part of France and Spain formed a distinct clade B, which, except for one isolate, had identical sequences in the analysed region (Fig. 3). This could indicate a recent introduction from a common source or a high level of evolutionary adaptation. Interestingly, viruses isolated more than 3000 km apart clustered within this group, which is surprising as the gC gene of PrV was shown to be variable even in epidemiologically linked outbreaks in domestic pigs [Reference Müller36]. Contamination as a reason for the observed clusters is highly unlikely as DNA was extracted at different locations and also independent PCRs were used for sequencing.
Further research is needed to elucidate transmission and spread of PrV in wild boar and feral pig populations. In contrast to clade B, clade A is more diverse comprising isolates from Slovakia, Hungary, eastern Germany, Italy and France. The reason for this difference relative to clade B remains elusive but may indicate a longer period of circulation with resulting diversification or multiple introductions. In any case, both clades seem to overlap geographically in central Europe, since, isolates of both groups were found in Germany and France.
Phylogenetic analysis suggests that PrV isolates of wild boar origin identified in Europe do not represent a homogenous population. If the diversity in the PrV isolates is taken into account, multiple introductions from domestic pigs into wildlife appear to be one plausible explanation for the diversity in the wild boar isolates with some occurring a very long time ago. This would correspond to observations made in France and Germany indicating that PrV infections were either not present in wild boar or had occurred only at a very low level [Reference Müller, Conraths and Hahn4, Reference Albina6]. On the other hand the virus may have been present already in wild boar in Europe and in wild pigs in the USA before, and may have caused several spillover infections at the time. Interestingly, in Germany, monitoring has provided evidence that PrV may have been circulating in wild boar for decades without confirmed spillovers to domestic pigs (T. Müller, unpublished data). Future studies to assess transmissions between wild boar and domestic pigs should include PrV isolates obtained from domestic swine from the same regions.
It was hypothesized that PrV in wild boar in Italy may have originated from the (illegal) movement of infected wild boars from eastern European countries to replenish the Italian wild boar population [Reference Capua20]. However, no information about PrV in wild boar in those countries was available at that time. In our analysis, the Italian isolate clusters within clade A. However, it is difficult to trace the origin of the individual PrV circulating in wild boar populations in Europe since molecular characterization uncovers genetic but not necessarily epidemiological relationships. This is particularly important if PrV isolates of wild boar origin which come from geographically distant regions are analysed. This also applies to the genetic homogeneity in clade B.
Considering the success of national Aujeszky's disease eradication programmes and the maintenance of an officially disease-free status of serveral countries, further knowledge on PrV infections in wildlife is increasingly important. Previous studies from Germany and Spain revealed no evidence for transmission of PrV between the domestic pig and the wild boar populations [Reference Lutz23, Reference Kükedi35, Reference Müller38, Reference Ruiz-Fons39]. Moreover, virus transmission from wild boar to domestic pigs has not been reported in Europe yet, although domestic pigs can be experimentally infected with PrV isolates originating from wild boar [Reference Müller22]. To consider a country or zone free from AD, the World Organization for Animal Health (OIE) requires that measures are implemented to prevent any transmission of PrV from wild boar to domestic pigs [40]. Therefore, the epidemiological situation in wild boar should be continuously monitored and assessed. In this respect, serosurveillance is an appropriate screening tool to verify the presence or absence of PrV. However, for more detailed information, virus isolation and subsequent molecular characterization is needed to trace possible virus transmissions. Our study contributes to these requirements as the obtained sequences will help in the trace-back process in the case of an emergency.
ACKNOWLEDGEMENTS
The authors thank the numerous hunters, technical staff and other helpers for their support in collecting and processing samples during targeted virological surveys in wild boar implemented in the respective countries. We are especially grateful to Doris Junghans and Diana Werner for their skillful technical assistance in propagating and characterizing the PrV isolates.
DECLARATION OF INTEREST
None.








