Introduction
Typhoid fever is caused by Salmonella enterica serotype Typhi. Salmonella Typhi is transmitted by ingestion of food and water contaminated by the faeces or urine of infected persons, as well as direct person-to-person transmission [Reference Harris, Ryan, Bennett, Dolin and Blaser1, Reference Heyman2]. Symptoms of typhoid fever are non-specific, and the incubation period ranges from 3 to more than 60 days, with a typical range of 8–14 days. Up to 5% of patients with acute typhoid fever become chronic carriers and can serve as a reservoir, enabling them to pass the disease to others [Reference Harris, Ryan, Bennett, Dolin and Blaser1, Reference Heyman2]. Typhoid fever is endemic in developing nations, affecting an estimated 20 million people worldwide and causing 200 000 deaths annually [Reference Harris, Ryan, Bennett, Dolin and Blaser1, Reference Crump, Luby and Mintz3–Reference Buckle, Walker and Black5].
Approximately 300 cases of typhoid fever occur in the USA each year, with more than 75% due to international travel [Reference Adams6–8]. Despite the low incidence of typhoid fever in the USA, outbreaks continue to occur due to contamination of food and water sources by infected individuals, or importation of contaminated products [Reference Olsen9–Reference Hancock-Allen16]. From 1960 to 1999, 54 typhoid fever outbreaks occurred in the USA. In 21 (39%) of these outbreaks, an asymptomatic carrier was identified as the source; 13 (62%) involved a carrier that was either foreign born, reported international travel within the last year, or had a history of typhoid fever [Reference Olsen9]. Of the 28 outbreaks detected from 2000 to 2010, 16 (57%) were attributed to a confirmed or suspected carrier [Reference Imanishi10].
On 2 February 2015, a case of typhoid fever was reported to the Oklahoma State Department of Health (OSDH) in a Marshallese resident of Kay County. The case-patient denied international travel during the 60 days prior to illness onset, but reported attending a funeral and related events during the weekend of 17–18 January 2015. The funeral and affiliated events occurred in Garfield County, Oklahoma; attendees included residents of several Marshallese communities in Oklahoma and other states, as well as guests from the Republic of the Marshall Islands (RMI). Over the next several days, additional cases of typhoid fever were reported, including in a Marshallese resident in Iowa that reported travelling to Garfield County, Oklahoma during the same weekend of the aforementioned funeral events and identified confirmed cases as contacts. The OSDH, in collaboration with the Centers for Disease Control and Prevention (CDC), conducted an outbreak investigation to identify the potential source, routes of exposure and to implement control measures.
Methods
Case definition
A confirmed case was defined as a person with isolation of S. Typhi from a stool or blood culture, with one of the designated outbreak patterns (XbaI restriction enzyme patterns JPPX01.0255, JPPX01.1200, JPPX01.1201 or JPPX01.1029) as determined by pulsed-field gel electrophoresis (PFGE), with illness onset from 1 January through 31 March 2015. A probable case was a person with clinically compatible illness (e.g. diarrhoea, fever, etc.) who had close, personal contact to a confirmed case. Close, personal contact was defined as either household contacts or individuals who attended any of the community gatherings linked to the outbreak.
Epidemiological investigation
We conducted active case finding by distributing a health alert network advisory to clinicians, acute care facilities and laboratories statewide, requesting cases to be reported immediately to OSDH for investigation. PulseNet, the national molecular subtyping network for enteric disease surveillance, was queried to identify S. Typhi cases with a PFGE pattern indistinguishable from one of the outbreak-associated strains.
In-person interviews were completed by OSDH epidemiologists and local county health department nurses using an outbreak-specific questionnaire. This questionnaire collected information on case-patient demographics; clinical history, including prior history of typhoid fever; and occupation to identify persons affiliated with high-risk settings for S. Typhi transmission, including patient care, foodservice, childcare and school. We attempted to identify the potential outbreak source by gathering exposure histories, including travel to the RMI, attendance of the funeral events and other group gatherings, consumption of foods imported from the RMI, hosting travellers visiting from RMI and other states, and contact with persons experiencing symptoms of acute enteric fever. We constructed social networks to identify common households, relatives, friends and work settings. We interviewed household contacts of cases regarding history of typhoid fever symptoms since January 2015.
Laboratory
Hospital laboratories forwarded S. Typhi isolates to the OSDH Public Health Laboratory (PHL) for confirmation and PFGE subtyping. The PHL confirmed Salmonella species using Biolog's GEN III MicroPlate™ test panel and Microbial Identification System software (Biolog Inc., Hayward, CA). Confirmed S. enterica isolates were further characterised using commercially available antisera (Thermo Scientific™/Denka Seiken™, San Jose, CA, USA) to detect O, Vi and H antigens present in S. Typhi. Local and state public health laboratories receive clinical Salmonella isolates for molecular subtyping and perform PFGE on all S. Typhi isolates according to PulseNet standard operating procedures for Salmonella, which includes digesting the DNA with restriction enzymes XbaI and BlnI [Reference Ribot17]. These public health laboratories submit subtyping results to PulseNet's national database, which is hosted and curated by database managers at CDC. PFGE patterns were analysed and evaluated to identify potential outbreak-associated isolates using Bionumerics version 6.6 software (Applied Maths/bioMérieux, Austin, TX, USA ).
Whole genome sequencing (WGS) was performed by CDC and the OSDH PHL to further characterise the level of genetic relatedness [Reference Sabat18]. Sequencing was performed on the Illumina MiSeq using NexteraXT (Illumina Inc., San Diego, CA, USA) library preparations and 2 × 250 bp sequencing chemistry. High-quality single-nucleotide polymorphism (hqSNP) analysis was conducted with Lyve-Set 1.1.4f (https://github.com/lskatz/lyve-SET) using an internal draft assembly (64 contigs) as reference with and without prophage masking [Reference Katz19]. Sequence results of outbreak isolates were evaluated in relation to the WGS profiles of approximately 2000 S. Typhi isolates utilised by Wong et al., to develop a database of four primary clusters, 16 clades and 49 sub-clades to characterise the likely geographic origin of typhoid fever cases [Reference Wong20].
Isolates were sent to CDC for antimicrobial susceptibility testing using broth microdilution (Sensititre®, Trek Diagnostics, part of Thermo Fisher Scientific, Cleveland, OH, USA) in the National Antimicrobial Resistance Monitoring System laboratory at CDC. The following antimicrobial agents were tested: amoxicillin–clavulanic acid, ampicillin, azithromycin, cefoxitin, ceftiofur, ceftriaxone, chloramphenicol, ciprofloxacin, gentamicin, nalidixic acid, streptomycin, sulfisoxazole, tetracycline and trimethoprim–sulfamethoxazole. Clinical Laboratory Standards Institute criteria were used for interpretation when available [21].
Intervention
We conducted community outreach with the goal to provide education about typhoid fever, and how to control further transmission. OSDH epidemiologists and local county health department nurses conducted lectures during Marshallese church meetings. Infographics and letters were developed in local Marshallese dialects and distributed to teach proper hand hygiene, discuss the importance of completing antibiotic therapy and instruct stool specimen collection. We also offered testing and treatment to symptomatic persons through local county health departments.
Confirmed and probable case-patients were restricted from high-risk settings, defined as attending or working in school, patient care, foodservice and childcare settings until two (for school settings) or three stool specimens, collected at a minimum of 30 days after symptom onset, were negative for S. Typhi. Case-patients were advised to collect the first specimen at least 48 h after completion of antibiotic therapy; subsequent specimens were collected at least 24 h apart. Stool collection kits with Cary Blair media were provided with instructions to return to the local county health department, or to schedule a home visit with an investigator for pick-up. We communicated directly with employers and institutions to ensure case-patients were excluded until testing indicated the individual was not shedding the organism.
Statistical analysis
Data were entered into a Microsoft Access 2010 database and analysed using SAS 9.4 software (SAS Institute, Cary, NC, USA). Analyses included confirmed and probable cases.
Results
Epidemiological investigation
We identified 38 cases of typhoid fever (Table 1); 25 (66%) were culture-confirmed S. Typhi and 13 (34%) were symptomatic contacts who met the probable case definition. Twelve (38%) cases were identified from a blood culture, nine (36%) from stool and four (16%) from both blood and stool. Of the 38 cases, 36 (95%) were Oklahoma residents and two (5%) were Iowa residents. Twenty-two (58%) cases were male. Thirty-seven (98%) reported their racial background as Native Hawaiian and other Pacific Islanders (NHPI); the remaining confirmed case-patient reported their race as Caucasian. The overall median age was 16 years (range: 1–57 years); 23 (61%) cases were children <18 years old.
Table 1. Demographic characteristics of case-patients by case status, typhoid fever outbreak investigation, Oklahoma, 2015

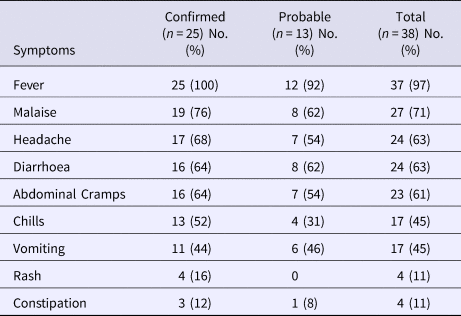
The date of symptom onset ranged from 19 January through 30 March 2015; the majority of cases occurred from 19 January through 23 February (Fig. 1). The most frequently reported symptoms included fever (97%), malaise (71%), headache (63%), diarrhoea (63%) and abdominal cramps (61%) (Table 2). Fourteen (37%) case-patients were hospitalised; all were laboratory-confirmed cases. The number of hospitalisation days ranged from 3 to 17 (median: 7 days). No deaths occurred due to this outbreak.

Fig. 1. Frequency of symptom onset by case status, typhoid fever outbreak investigation, Oklahoma, 2015.
Table 2. Clinical summary of case-patients by case status, typhoid fever outbreak investigation, Oklahoma, 2015
We were unable to identify a specific source of this outbreak. Thirty-five (97%) of 36 Oklahoma cases were clustered among an extended network of relatives, household members and acquaintances. Twenty (52%) case-patients reported attending the funeral and associated social events during early January 2015; however, they chose to not provide information regarding foods served and other relatives or acquaintances in attendance. The confirmed case-patient with the earliest onset date of 19 January informed investigators that a relative from another state was experiencing symptoms of acute illness while attending the funeral events and prepared food for the service; however, we were unable to identify this symptomatic contact, since relatives declined to provide identifying information. Another confirmed case-patient reported consuming fish, coconut and beru that were brought to Oklahoma by relatives from the RMI. Several case-patients acknowledged persons from the RMI often bring traditional foods when they visit, but declined to provide details regarding consumption of specific foods imported prior to illness onset, attendance of Marshallese social events and foods served during group events that were locally prepared or imported by attendees. Both Iowa residents reported travel to Oklahoma during their exposure period to visit relatives, and reported attending the funeral events as well as identified several Oklahoma case-patients as contacts. The one Caucasian case-patient (illness onset date 30 March) denied attending any Marshallese events, but was a resident of Kay County where several outbreak-associated cases resided. Although an extensive exposure history was collected from the case-patient, we were unable to identify an epidemiological linkage to another case or common exposure.
Laboratory
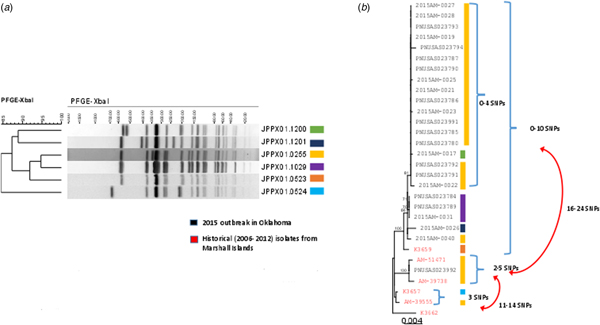
Isolates for all 25 confirmed cases were submitted to the Public Health Laboratories in Oklahoma and Iowa. PFGE analysis identified four closely related patterns (Fig. 2a). Isolates from 18 (72%) cases had the primary outbreak pattern JPPX01.0255; the four other patterns consisted of one to four cases each (Fig. 2a). PFGE patterns were uploaded to the PulseNet national database and compared with other S. Typhi patterns. No other outbreak-associated cases were identified. Twenty isolates were submitted to CDC for susceptibility testing; all were susceptible to all antimicrobial agents tested.
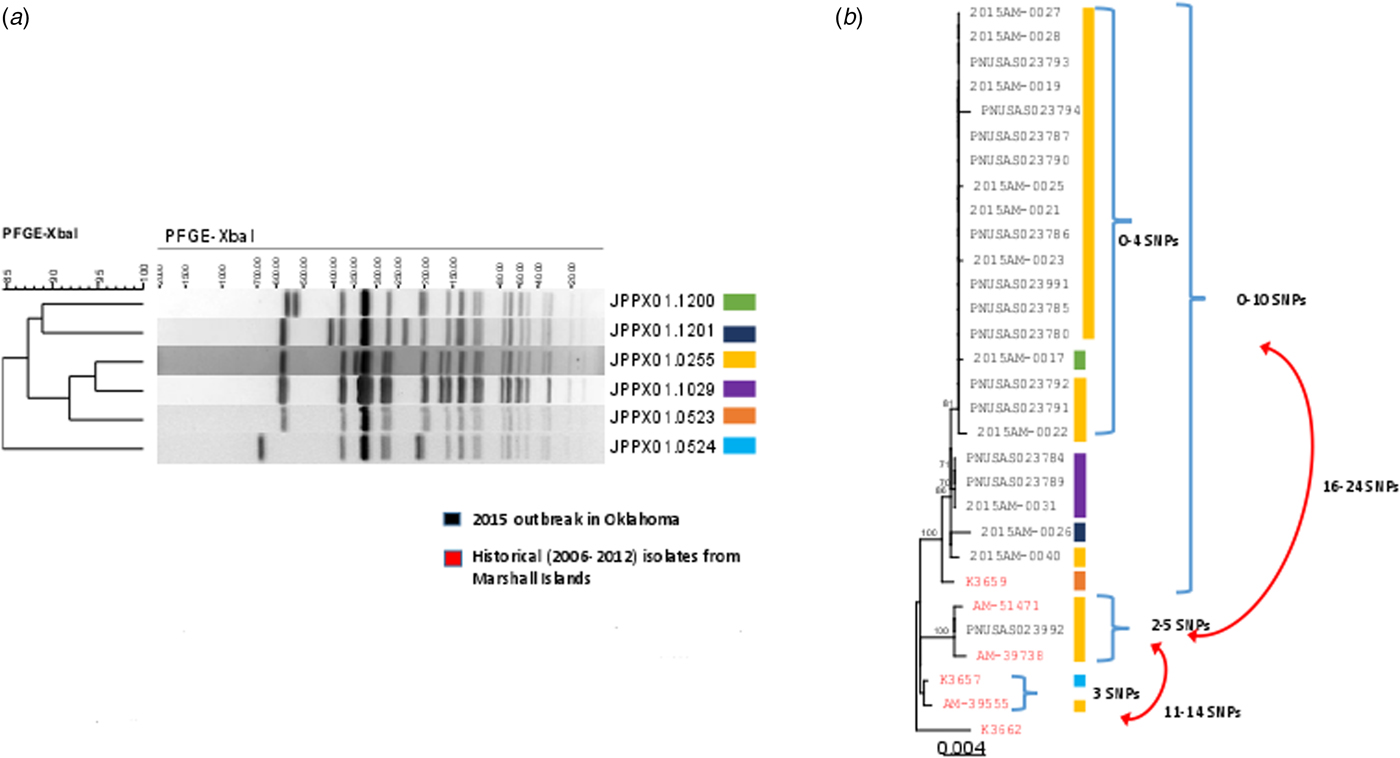
Fig. 2. PFGE patterns (JPPX01.1200, JPPX01.1201, JPPX01.0255, JPPX01.0523, JPPX01.0524) and phylogenetic tree of Salmonella Typhi outbreak isolates associated with the typhoid fever outbreak investigation, Oklahoma, 2015. (a) Four different XbaI PFGE patterns identified among the 24 clinical isolates associated with the outbreak (green, navy blue, yellow and purple) and two PFGE patterns detected among the six historical Salmonella Typhi isolates with a connection to Marshall Islands (orange and blue). (b) Phylogenetic tree based on the hqSNP analysis for the 24 clinical isolates included in the 2015 outbreak in Oklahoma (black font) and six historical isolates with a connection to Marshall Islands (red font). Reads were trimmed before mapping by SMALT [Reference Katz19]. SNPs were called using Varscan at >20× coverage, >95% read support and <5 bp apart. Draft assembly K3657 (SPAdes-3.6.1) was used as a reference without phage masking. Six different PFGE patterns were detected among the 30 isolates (colour-coded bars).
A total of 24 isolates from the outbreak were included in the WGS analysis. In addition, three historical isolates collected from case-patients with history of travel to the Marshall Islands, and three isolates from a 2006 Marshall Islands S. Typhi investigation were included for comparison. WGS revealed 23 of the 24 clinical isolates (22 Oklahoma, one Iowa), and one historical isolate from 2006 were genetically highly related clustering within 0–10 single-nucleotide polymorphisms (SNPs) (Fig. 2b). The second isolate from Iowa clustered with two historical isolates: one from a 2009 Oklahoma case and another from a 2012 Hawaii case with travel history to the Marshall Islands; however, these isolates differed from the Oklahoma outbreak clade by 16–24 SNPs. Prophage masking did not have an effect on the SNP counts.
When compared with the extended genotyping framework of S. Typhi isolates developed by Wong et al., Oklahoma and Iowa outbreak isolates all fit into subclade 2.3.3 with at least 96% certainty. The likely countries of origin for isolates utilised by Wong et al. that fit into subclade 2.3.3 include South America, South Asia and Southeast Asia. South American cases included in their analysis were associated with travel to Bangladesh [Reference Wong20].
Intervention
OSDH epidemiologists and local county health department nurses conducted seven lectures at Marshallese church meetings, and delivered information on typhoid fever to members of each congregation, as well as to household members and acquaintances during home visits. We collaborated with Garfield County school officials and businesses that were known to be common employers of Marshallese residents, including a food manufacturing plant and home health services, to establish contacts to report symptomatic employees with symptoms suggestive of typhoid fever. Sixteen (42%) cases, four confirmed and 12 probable, were identified through these outreach events. An additional 207 household members and acquaintances were identified as contacts to cases and interviewed during home visits to case residences.
Twenty-seven (71%) cases were affiliated with a high-risk setting; 22 (82%) attended school, three (11%) worked at a manufacturing plant involving direct handling of food products and two (7%) worked as direct patient care providers (Table 3). OSDH epidemiologists and local health department nurses notified school officials and employers of persons that needed to be excluded due to the public health investigation.
Table 3. High-risk setting affiliation by case status and completion of stool submission for exclusion requirements, typhoid fever outbreak investigation, Oklahoma, 2015

a One Iowa case-patient was affiliated with a school setting; however, exclusion and stool submission was not required. Exclusion and testing for cases affiliated with schools were only implemented for Oklahoma residents.
Of the 27 cases affiliated with high-risk settings, 23 (85%) submitted the required stool specimens to confirm they were no longer shedding the organism. All five patient care providers and food handlers complied with the requirement of submitting three specimens that were negative for S. Typhi in order to return to work. The median timeframe of date of typhoid fever case report to exclusion from high-risk setting was 2 days (range: 0–19 days). Three cases were excluded from school, but were lost to follow-up on attempts to collect stool specimens. Of those that completed follow-up stool cultures, date of exclusion to final negative stool culture result ranged from 27 to 204 days, with a median duration of 60 days. The extended timeframe of exclusion was partly due to the decision by several parents of case-patients to delay their children's stool sample submission until after the summer break. Exclusion and stool submission requirements were not implemented for the one school-affiliated Iowa case, because this control measure was Oklahoma-specific.
Discussion
This is the largest outbreak of typhoid fever in the USA in the last 27 years. The previous largest typhoid outbreak in the USA was associated with consumption of contaminated orange juice served at a hotel; this outbreak involved 67 cases total, including 43 culture-confirmed and 24 probable cases among hotel guests, one culture-confirmed case and one asymptomatic culture-positive case among hotel employees, and one culture-confirmed secondary case [Reference Birkhead22].
Although we did not identify a vehicle, this outbreak of typhoid fever among Marshallese residents of Oklahoma and Iowa likely resulted from consumption of contaminated food served during the funeral or associated social event, and person-to-person transmission among a network of relatives, household members and close acquaintances. This is supported by the temporality of cases with a common history of attending the same funeral events in Oklahoma, the occurrence of disease among an extended network of relatives, household members and acquaintances, and WGS results that revealed S. Typhi isolates representing the four PFGE strains identified in this outbreak were highly genetically related.
The primary outbreak-associated PFGE pattern in this outbreak of typhoid fever has historically been associated with travel to the RMI. From 2009 to 2014, seven isolates with this pattern were from Hawaii (n = 6) and Pennsylvania (n = 1); all case-patients reported a history of travel to the RMI. Similar to cases from this outbreak, none of the previous cases with travel to the RMI demonstrated evidence of antimicrobial resistance. Public health officials in RMI were contacted during this outbreak to ensure that there was not a concurrent outbreak going on in-country; no outbreak was occurring and the last report of any typhoid fever case to the RMI health department was in October 2014.
Typhoid fever is endemic in the RMI [Reference Crump, Luby and Mintz3]. The Compact of Free Association Act of 1985 established the RMI as an independent nation, and established a relationship between the USA and RMI that allows citizens to apply for admission to live and work in the USA as non-immigrants without visas [23]. The Marshallese population has rapidly grown representing more than 22 000 of the NHPI that reside in the USA. According to the 2010 Census, approximately 2000 Marshallese reside in Garfield County, Oklahoma, which ranked 10th for counties with the highest proportion of NHPI residents [Reference Hixson, Hepler and Kim24]. This outbreak involved residents of both Garfield and Kay counties in Oklahoma, highlighting how a non-endemic disease such as typhoid fever can spread from an endemic area (in this case, the RMI) to specific communities within the USA.
This investigation included the first application of WGS in the USA to complement interpretation of epidemiological and laboratory results during an outbreak of typhoid fever comprised of multiple PFGE strains. WGS provides a more refined molecular characterisation of the genetic relatedness between isolates than PFGE. WGS revealed a 0–10 SNP variation between most of the isolates that represented the four outbreak-associated PFGE patterns; this indicates a very close genetic relationship among the isolates, suggesting that they may have been from a common source. Furthermore, testing of historical isolates of typhoid fever cases among persons with a history of travel to the RMI indicated a close genetic relatedness. The three isolates that differed from the OK outbreak clade may indicate that this outbreak was polyclonal. This hypothesis is supported by reports from cases that relatives and members of the RMI community were involved in the preparation of food for the funeral events, and several travellers brought foods from the RMI for relatives, which could have led to the introduction of and exposure to more than one strain of S. Typhi.
Four different PFGE patterns were included in this outbreak. It is not unusual to have multiple PFGE strains in a single S. Typhi outbreak; a single individual or chronic carrier can shed multiple genetically distinct variants [Reference Imanishi10]. Although we were able to link all case-patients to other probable or laboratory-confirmed cases, the identification of multiple PFGE patterns, the inability to implicate a common source and knowledge of travellers from the RMI and other states limited our ability to rule out the potential that some case-patients were secondary cases to more than one symptomatic or chronic carrier.
The results of this investigation illustrate that implementation of control measures limited continued transmission in high-risk settings. We interacted directly with schools, foodservice establishments and patient care settings to assure timely exclusion of cases until follow-up testing requirements were met. No secondary cases were identified in institutional settings. Only one case was identified in a person who did not report a history of attending the Marshallese funeral events or was a close, personal contact to a confirmed or probable case.
There were significant challenges encountered during this outbreak investigation. We were unable to conduct an epidemiological study to identify the source, since Marshallese residents were hesitant to answer questions regarding the funeral events under investigation, including foods served and names or contact information of attendees. This limited our investigation to a descriptive analysis of cases and contacts rather than an analytic study to measure associations between exposures and outcomes. Although over 200 household members were identified and interviewed during the contact investigation, Marshallese residents were reluctant to provide information regarding other acquaintances. Several case-patients acknowledged that funeral attendees included travellers from other states and the RMI, but would not share identifying information. Although we conducted meetings with local Marshallese leaders to encourage participation and worked with interpreters to complete interviews, many individuals expressed reluctance about participating for fear of stigmatizing Marshallese residents by association with an outbreak. Finally, delays resulted from the need for interpreters as many Marshallese residents required interpretation for interviews. In spite of these limitations, the absence of clusters of disease within high-risk settings or multiple cases with no clear epidemiological linkage to the events under investigation demonstrate we were successful in limiting person-to-person transmission.
This investigation highlights the risk of typhoid fever among communities comprised of international populations from endemic countries. Public health officials and clinicians should consider imported diseases in their differential diagnosis in persons with close ties to endemic areas in the absence of international travel history or known exposure to a symptomatic contact. Additionally, WGS may be used when available to provide information on the genetic relatedness of isolates in typhoid fever outbreaks. Lastly, timely implementation of control measures is paramount to limiting sustained transmission of typhoid fever in high-risk settings by outbreak-associated cases.
Acknowledgment
The authors sincerely thank the numerous employees of the Oklahoma State Department of Health Acute Disease Service, Public Health Laboratory, Garfield County Health Department, and Kay County Health Department that contributed to this outbreak investigation. We also thank the Iowa Department of Public Health Center for Acute Disease Epidemiology and State Hygienic Laboratory for their case-patient interviews and specimen testing, and Michael Judd, MPH and L. Hannah Gould, PhD at the Centers for Disease Control and Prevention for their assistance with surveillance. We would like to acknowledge essential partners that assisted with case-patient finding, outreach, and control measures including the faith-based organizations that serve the Marshallese community in Garfield County; Debra Bartel, RN, Joe Snodgrass, MD, and Jarrod Mueggenborg, DO of St. Mary's Regional Medical Center; Joan McIntyre, RN, BSN of Enid Public Schools; and Earl Johnson of AdvancePierre Foods.
Financial support
None.
Conflict of interest
None.