INTRODUCTION
Rotaviruses are the chief aetiological agent of viral gastroenteritis in infants and young children, and in the young of a large variety of animal species [Reference Desselberger, Gray and Desselberger1, Reference Kapikian, Hoshino, Chanock, Knipe and Howley2]. In developing countries, it is estimated that rotavirus is responsible for one third of all diarrhoea-associated hospitalizations and between 450 000 and 650 000 infant deaths per year [Reference Bresee3].
Two rotavirus proteins, VP7 and VP4, which constitute the outer layer and the viral spikes, respectively, are known to induce neutralizing antibody responses and, therefore, protection from infection and disease is believed to be type-specific [Reference Desselberger, Gray and Desselberger1]. The classification of rotaviruses into G- and P-types is based on the variability observed in the genes encoding these two proteins. At least 15 G-types (G1–G15) and 20 P-types (P[1]–P[20]) have been identified to date, of which 10 G-types and five P-types have been found in rotaviruses infecting humans [Reference Bresee3].
The middle layer of rotaviruses is constituted by a trimeric protein, VP6, that interacts with the inner-layer protein VP2 and with the two outer-layer proteins VP7 and VP4 [Reference Desselberger, Gray, Estes, Mahy and Ter4]. Among group A rotaviruses, four different subgroups (SG), SGI, SGII, SGI+II and SGnonI/nonII have been distinguished on the basis of VP6 diversity, but human rotavirus strains are predominantly of SGI or SGII [Reference Iturriza-Gomara5].
The segmented nature of the rotavirus dsRNA genome allows reassortment between strains, and the extensive genome diversity resulting from the accumulation of point mutations results in co-circulating rotavirus strains with various combinations of VP7, VP4 and VP6 each of which can be selected from multiple genetic lineages.
In this study, we describe the detailed characterization of rotavirus strains co-circulating in Tehran, Iran, between November 2003 and October 2004. The distribution of G-types, P-types and subgroups was analysed by molecular methods, including reverse transcriptase–polymerase chain reaction (RT–PCR), semi-nested RT–PCR and nucleic acid sequencing.
METHODS
Clinical specimens
A total of 374 faecal samples, collected from children aged 6 months to 5 years, suffering acute diarrhoea of unknown origin, were tested for rotavirus antigen by ELISA (IDEIA Rotavirus, Dako, Denmark). The assay was performed according to the manufacturer's instructions. The children were either in-patients (admitted to hospital) or outpatients (visited the emergency room) of the Markaz Tebbei Koudakan (Children's) Hospital in Tehran, Iran, from November 2003 to October 2004. The study paediatrician assessed the severity of disease by examining the child and interviewing the mother. A point scale, based on the frequency and severity of diarrhoea (bloody, mucus, watery), episodes of vomiting, associated fever (<38·5°C, >38·5°C) and dehydration (mild, moderate, severe), was used to record the severity of illness [Reference Ruuska and Vesikari6]. Data on age, sex, nutrition (breast milk, milk formula) were also collected.
Stool samples were stored at 4°C and a subset of 92 samples was sent to the Enteric Virus Unit, Centre for Infections, HPA, London, UK, for rotavirus characterization.
Nucleic acid extraction and G- and P-typing, RT–PCR
Faecal suspensions (10%) in balanced salt solution 199 were used for nucleic acid extraction using the guanidinium isothiocyanate/silica method [Reference Boom7]. The cell culture-derived reference strain SA11 was used as a positive control and RNase-free distilled water used as a negative control throughout. Reverse transcription in the presence of random primers, G-typing, P- typing, and subgroup PCRs were performed as described previously [Reference Gentsch8–Reference Iturriza-Gomara, Kang and Gray10]. PCRs were performed as described previously [Reference Iturriza-Gomara, Kang and Gray10]. Amplicons were detected by electrophoresis using a 3% NuSieve 3:1 (Cambrex, Nottingham, UK) agarose gel in 1× TAE buffer and electrophoresed at 7–8 V/cm for 75–90 min, and stained with ethidium bromide (0·5 μg/ml) and photographed using a Bio-Rad GelDoc system (Bio-Rad, Hemel Hempstead, UK).
Rotaviruses that failed to genotype
Amplicons of 881 bp of the gene encoding VP7 from nine rotavirus-positive specimens, that failed to G-type, were sequenced using VP7 consensus primers VP7-F and VP7-R [Reference Iturriza-Gomara, Kang and Gray10]. A VP7 nested consensus RT–PCR that amplified a 293-bp fragment was used for the remaining 11 rotavirus-positive samples, which had no bands detectable after the first round of G-typing RT–PCR. The forward primer was VP7-F and the reverse primer was VP7-RINT: ANAYNGANCCWGTYGGCCA (nt 331–344), and the PCR conditions were as previously described for first round G-typing PCR, but with primer annealing performed at 50°C [Reference Iturriza-Gomara, Kang and Gray10]. The first-round PCR products of G-typing reactions and nested PCR products were purified according to the manufacturer's instructions using a Geneclean® kit (Anachem, Luton, UK), and sequenced using a Beckman Coulter™ CEQ2000 Dye Terminator Cycle Sequencing Quick Start kit (according to manufacturer's instructions, Beckman Coulter, High Wycombe, UK) and a Beckman Coulter CEQ2000 capillary sequencer (Beckman Coulter). The sequences were compared against sequences in a database containing a collection of over 400 sequences of genes encoding VP7, VP4 and VP6. Data analysis was performed using Clustal for multiple alignments and neighbour joining for phylogenetic analysis (Bionumerics, Kortrijk, Belgium). Sequences were deposited in the Rotavirus typing database at the Centre for Infections, HPA, London, and are feely available from the corresponding author.
Statistical methods
The significance of differences was determined using the χ2 test.
RESULTS
Rotavirus antigen detection
Rotavirus antigen was detected by ELISA in 92 out of 374 (24·6%) samples. A total of 64 ELISA-positive samples and 28 ELISA-negative samples were sent to the Enteric Virus Unit, Centre for Infections, London for further characterization.
Clinical and epidemiological analysis
Rotavirus infection was found in all age groups tested but was highest in children aged 18–20 months (Fig. 1). A significant association between fever, vomiting, diarrhoea and a negative association with breastfeeding were sought. Vomiting (P<0·0001), diarrhoea (P=0·0001), and severe dehydration (P<0·0001) were significantly associated with rotavirus infection (data not shown). Children who were breastfed exclusively or in conjunction with formula feeds were less likely to have rotavirus infection when compared to children fed exclusively on milk formula, but this difference was not statistically significant. There was no clear temporal distribution of rotavirus infections although peaks were detected in the spring, late summer and autumn (Fig. 2).
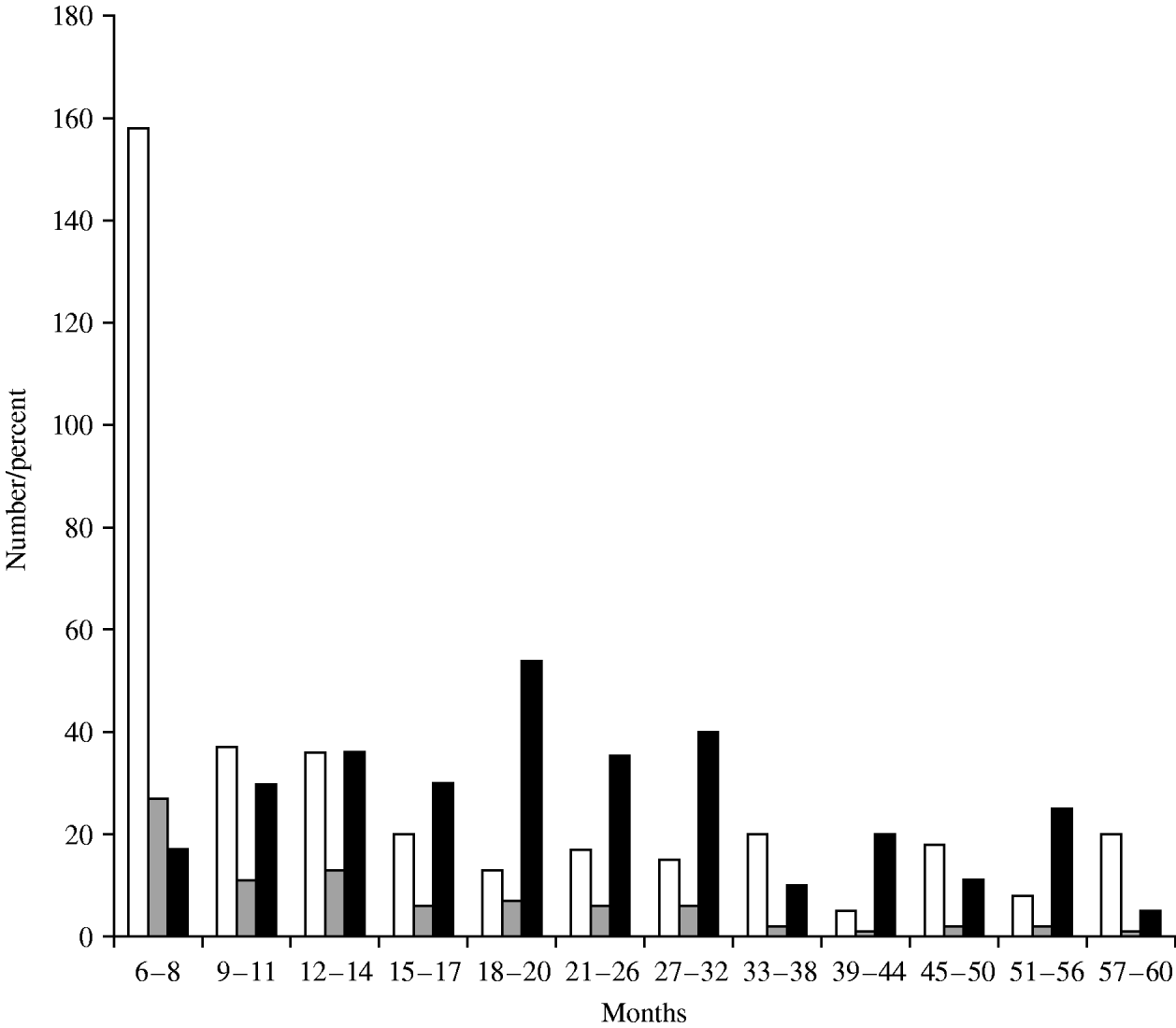
Fig. 1. Age distribution of rotavirus infection. □, Total;
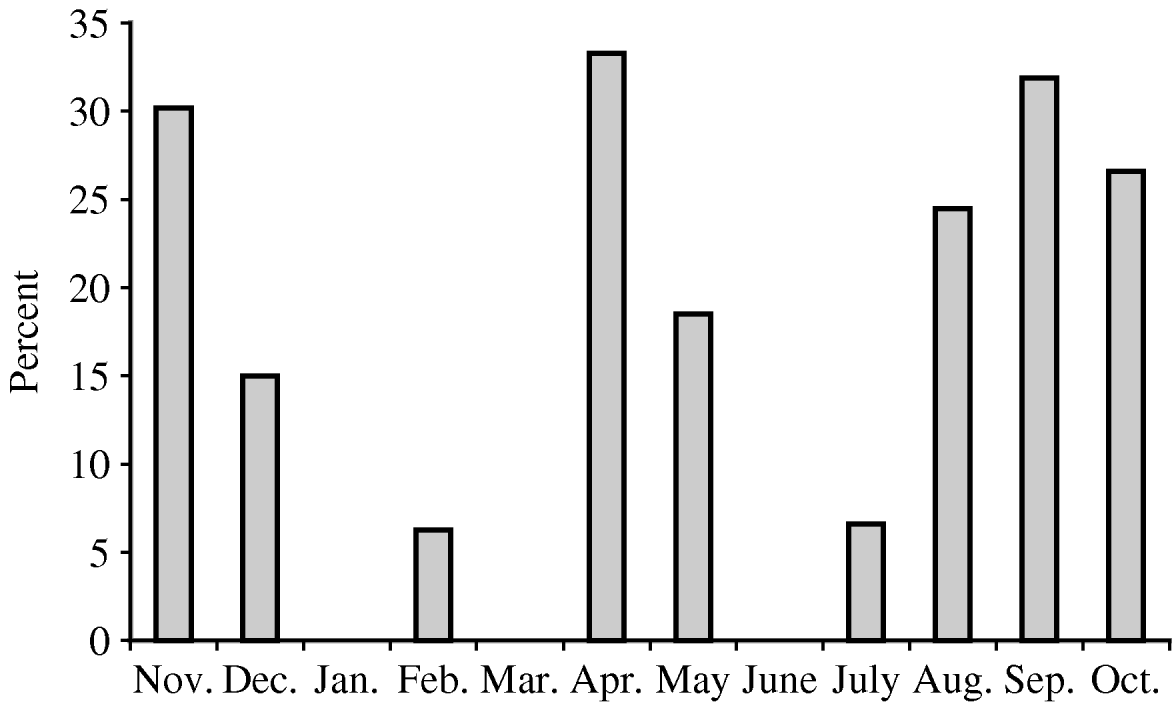
Fig. 2. Temporal distribution of rotavirus infections.
Rotavirus characterization
Of the 64 rotavirus ELISA-positive samples, three were negative in VP7-, VP4- and VP6-specific RT–PCRs and of the 28 ELISA-negative samples, rotavirus RNA was detected by VP6 RT–PCR in 10.
Genotyping to determine the G-type, P-type and subgroup was performed on 71 samples. Rotaviruses of G1, G2, G4, G9, G12 and P[4], P[8] and P[10] were found and G1P[8] SGII (59·2%) and G9P[8] SGII (15·5%) were the predominant rotavirus genotype combinations detected (Table). Mixed rotavirus infections were detected in samples collected from two children and the G- and P-types were not determined in six samples and one sample, respectively. No significant differences in genotype distribution between strains collected from hospital in-patients and outpatients were detected.
Table. Rotavirus G, P and SG combinations co-circulating in Tehran, Iran
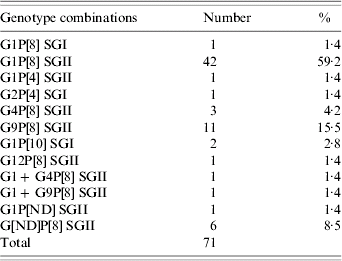
ND, Not detected.
DISCUSSION
Rotavirus is an important cause of childhood morbidity in developed and developing countries, and of mortality in developing countries. In Iran, the mortality rate in children aged <5 years is 64/1000. A total of 10 880 deaths per year in children are associated with diarrhoeal illness and it is estimated that rotavirus infection is responsible for more than 2700 deaths per year (M. Taremi, M. R. Zali, personal communication).
Studies in Iran, conducted over the last 5 years, reported that rotavirus was detected in 15·3–26·3% of children with gastroenteritis and the highest incidence of infection was in children aged 6–12 months [Reference Zarnani11, Reference Samarbafzadeh12]. Zarnani et al. [Reference Zarnani11], described a seasonal incidence with infections peaking in the spring, whereas in our study we found two peaks, one in spring and another in autumn, however, more extensive data from consecutive years is necessary in order to reach any firm conclusion regarding the seasonality of rotavirus infections in Iran. In our study, rotavirus was detected in 24·6% of children with gastroenteritis and the highest incidence of infection was in children aged 18–20 months. A previous study conducted in Zahedan city, Iran, reported the highest incidence in hospitalized children was between the ages of 19 and 24 months [Reference Moradi and Mokhtari-Azad13]. The reason for this shift to a higher incidence in older children is unclear but may be associated with prolonged breastfeeding or changes in the population of viruses co-circulating in Iran. No clear seasonal peak was determined in this study although more cases were seen in spring and late summer/early autumn.
Although significant associations were detected between rotavirus infection and diarrhoea, vomiting and severe gastroenteritis in our study, and the study of Zarnani et al. [Reference Zarnani11], we were unable to confirm that rotavirus infection was significantly less frequent in children exclusively breastfed. This may reflect the differences in peak age of rotavirus infection reported in the two studies.
The advent of monovalent and multivalent rotavirus vaccines offering homotypic protection against severe disease caused by commonly co-circulating rotavirus strains has been the driver to develop national and regional rotavirus surveillance schemes. The efficacy of vaccines within a particular population may be dependent on the rotavirus genotypes co-circulating in that population.
In a recent genotyping study conducted in Iran in 2001–2002, G1P[8], and G2P[4] were found to be the most common rotavirus genotype combinations circulating [Reference Moradi and Mokhtari-Azad13, Reference Khalili14]. In this study, both common and uncommon rotavirus genotypes were detected. Although G1P[8] SGII rotaviruses were the most commonly found, G9P[8] SGII not previously reported from Iran, was the second most commonly circulating strain. Unusual genotypes, G1P[10] SGI and G12P[8] SGII and strains derived from reassortment between common co-circulating genotypes such as G1P[4] SGII represented 5·6% of strains characterized.
G-typing was initially attempted in Tehran, Iran, using the methods and oligonucleotide primers described by Gouvea et al. [Reference Gouvea9]. The G-type was determined in 29 out of 49 (59·2%) samples tested and all were identified as G1.
Rotavirus G-typing in London, UK, employed a variety of methods using the primers described by Iturriza-Gomara et al. [Reference Iturriza-Gomara, Kang and Gray10]. A conventional genotype-specific multiplex semi-nested PCR assay was used initially and G-types were determined for 45 samples. A further nine samples were genotyped by sequencing the first-round PCR product of samples which failed to genotype with the second-round PCR genotype-specific primers. Through the use of a nested PCR which amplified a 293-bp region of the gene encoding VP7 a further 11 samples were genotyped following sequencing of the PCR amplicon. Therefore, using this sequential approach, the G-type was determined for 65 out of 71 (91·5%) samples.
The results of this study, and others from this region highlight the need for continued surveillance in conjunction with a testing regime that takes account of the rapid evolution of rotaviruses. Also, this study stresses the need to continually update genotyping assays to reflect the accumulation of point mutations at oligonucleotide primer binding sites and to detect novel strains emerging in the human population [Reference Iturriza-Gomara, Kang and Gray10].
ACKNOWLEDGEMENTS
The authors thank the staff of the Markaz Tebbei Koudakan (Children's) Hospital in Tehran and the WHO, Geneva, who provided a travel and training grant (ref. V27-370-1) to Dr Firouzeh Farahtaj to undertake this work at the Enteric Virus Unit, Virus Reference Department, Centre for Infections, Health Protection Agency, London, UK.
DECLARATION OF INTEREST
None.





