Introduction
Cytochrome P450 monooxygenases (P450s), is a super-family of heme-containing proteins, found in almost all living organisms from bacteria to human (Feyereisen, Reference Feyereisen2006). Cytochrome P450s constitute an important metabolic system and this system plays a central role in the oxidative metabolism of both xenobiotic and endogenous compounds (Feyereisen, Reference Feyereisen1999). In insects, they are best known for their roles in the metabolism of insecticides, which often results in the development of insecticide resistance (Zhou et al., Reference Zhou, Ma, Li, Sheng, Liu and Qiu2010). To date, more than 1000 cytochrome P450 genes in insects have been identified (Nelson, Reference Nelson2009), most of them are distributed to the families of microsomal CYP4, CYP6, CYP9, CYP28, CYP321 and mitochondrial CYP12 (Feyereisen, Reference Feyereisen, Gilbert, Iatrou and Gill2005). Furthermore, many cytochrome P450 genes, especially belonging to the families of CYP4, CYP6, CYP9 and CYP12, are frequently involved in insecticide metabolism and resistance (Feyereisen, Reference Feyereisen2006; Li et al., Reference Li, Schuler and Berenbaum2007).
It has often been mentioned that insecticide metabolism is enhanced by overexpression of cytochrome P450s and it appears to be a frequent type of resistance mechanism in insects (Liu & Scott, Reference Liu and Scott1998; Li et al., Reference Li, Yang, Wu, Li and Wu2006). Overexpression of cytochrome P450s has been reported in many insect species. For example, increased transcription of CYP6D1 (Liu & Scott, Reference Liu and Scott1998) in Musca domestica pyrethroid-resistant strain; CYP6BG1 and CYP6BG2 in permethrin-resistant strain of Plutella xylostella (Bautista et al., Reference Bautista, Tanaka and Miyata2007); CYP4H1, CYP4H22v1, CYP4J6v1 and CYP4J6v2 in deltamethrin-resistant strain of Culex pipiens pallens (Shen et al., Reference Shen, Dong, Tian, Ma, Li, Wu and Zhu2003). Expression profiles of cytochrome P450 genes are highly diverse in insects (Scott & Wen, Reference Scott and Wen2001). And different developmental stage or tissue-specific expression patterns may imply their specific functions, such as the metabolism of signal molecules, adaptation to host plants or insecticide resistance (Chung et al., Reference Chung, Sztal, Pasricha, Sridhar, Batterham and Daborn2009; Feyereisen, Reference Feyereisen and Gilbert2012). In insects, higher cytochrome P450s activities are usually associated with the midgut, fat bodies and Malpighian tubules (Brun et al., Reference Brun, Cuany, Le Mouel, Bere and Amichot1996). Inducibility is a general characteristic of cytochrome P450s, and it has been well documented in a number of insect species (Fuchs et al., Reference Fuchs, Spiegelman and Belitsky1994; Harrison et al., Reference Harrison, Zangerl, Schuler and Berenbaum2001). Several cytochrome P450 genes induced by xenobiotic, such as insecticides have been identified (Ranasinghe & Hobbs, Reference Ranasinghe and Hobbs1998; Brandt et al., Reference Brandt, Scharf, Pedra, Holmes, Dean, Kreitman and Pittendrigh2002; Poupardin et al., Reference Poupardin, Reynaud, Strode, Ranson, Vontas and David2008; Baek et al., Reference Baek, Clark and Lee2010). Xenobiotic inducible cytochrome P450 genes may be more likely to be involved in resistance (Le Goff et al., Reference Le Goff, Hilliou, Siegfried, Boundy, Wajnberg, Sofer, Audant, ffrench-Constant and Feyereisen2006). Therefore, it is important to study the expression profiles of cytochrome P450 genes and examine their inducibility by the particular insecticides, the findings may help researchers better understand functions of the genes and their interactions with insecticides at molecular levels and provide useful genetic information to assess potential consequences of insecticide exposures in insects.
The diamondback moth (DBM), P. xylostella, is a notorious insect and one of the most destructive cruciferous vegetables pests in the world due to its extreme ability to develop resistance (Talekar & Shelton, Reference Talekar and Shelton1993). Worldwide cost for chemical control of P. xylostella is about $4–5 billion (US dollar) annually (Furlong et al., Reference Furlong, Wright and Dosdall2013). Chlorantraniliprole, as an alternative to traditional insecticides, is a reduced-risk insecticide being introduced for global control of P. xylostella and has been shown to be a highly efficacious insecticide in pest control (Wang et al., Reference Wang, Li and Wu2010). However, due to intensive use of chlorantraniliprole, high levels of resistance in some field populations of P. xylostella have been identified in Guangdong province, China (Wang & Wu, Reference Wang and Wu2012). As an important detoxification enzyme family, cytochrome P450s play an important role in the resistance of P. xylostella to various insecticides, including chlorantraniliprole (Ninsin & Tanaka, Reference Ninsin and Tanaka2005; Wang et al., Reference Wang, Khakame, Ye, Yang and Wu2013). To date, ten cytochrome P450 genes have been identified from P. xylostella, among them, CYP6BG1 and CYP6BG2 are observed to be constitutively overexpressed in a permethrin-resistant strain (Bautista et al., Reference Bautista, Tanaka and Miyata2007), and the CYP6BG1 gene has been evaluated for its role in resistance by the RNA interference (RNAi) method (Bautista et al., Reference Bautista, Miyata, Miura and Tanaka2009). However, no P450 genes have been identified or reported to be responsible for chlorantraniliprole resistance in P. xylostella.
The previous studies on the transcriptome analysis of chlorantraniliprole resistance development in P. xylostella showed that the up-regulation of detoxification genes (P450, GSTs and prophenoloxidase) and the down-regulation of ryanodine receptors (the chlorantraniliprole-blinding target) are the main factors leading to the development of chlorantraniliprole resistance (Lin et al., Reference Lin, Jin, Hu, Chen, Yin, Li, Dong, Zhang, Ren and Feng2013). Here, based on one of the up-regulation cytochrome P450 genes cDNA fragments, a novel cytochrome P450 gene CYP321E1 was identified and reported for the first time. Furthermore, the gene expression profile, inducibility by three representative insecticides and its specific detoxification function using RNAi were also observed. Our findings will help elucidate its possible role in chlorantraniliprole resistance of P. xylostella and shed on new light on functional importance of this gene in detoxification of insecticides.
Materials and methods
Insects
The studies were carried out on a chlorantraniliprole-resistant strain of P. xylostella (GXA), as reported by Lin et al. (Reference Lin, Jin, Hu, Chen, Yin, Li, Dong, Zhang, Ren and Feng2013), which was collected from a cabbage field in Guangxi Province. The strain was maintained on cabbage in the laboratory for more than 20 generations, and selected using chlorantraniliprole every 2 to 3 generations to mitigate the influence of sensitive individuals and other field factors. The laboratory conditions were maintained at 25±2 °C, 70–80% relative humidity (RH) and 16:8 h light: dark photoperiod. Adults were fed on 10% honey solution and allowed to lay eggs on aluminum-foil paper with cabbage juice.
cDNA cloning of CYP321E1
Total RNA was extracted using Easy Spin Total RNA Extraction Kit (Biomed, China) and the first-strand cDNA was synthesized from 2 μg RNA with an oligo (dT) primer using Reverse Transcritase M-MLV (Takara, China) according to the manufacturer's instructions. The resulting cDNA was used as a PCR template with the following primers: 5′-GCT ACA ACC GGC TTT ATT CG-3′ and 5′-AAC AGG CAG GAT GGA TTC AC-3′. The primer designs were based on up-regulation cytochrome P450 gene cDNA fragments: CL6103.contig 1, which was obtained from P. xylostella EST database (Lin et al., Reference Lin, Jin, Hu, Chen, Yin, Li, Dong, Zhang, Ren and Feng2013). PCR was performed for 1 cycle at 94 °C for 5 min, then for 35 cycles at 94 °C for 35 s, 42 °C for 35 s and 72 °C for 1 min, followed by 1 cycle at 72 °C for 10 min. The amplified fragment was purified using a Gel Extraction Kit (Omega, China), then cloned into the pMD18-T vector (Takara, China) and sequenced. To get the full-length cDNA of CYP321E1, the 5′- and 3′-end specific primers were designed according to the manufacturer's protocol of SMART RACE cDNA Amplification Kit (Clontech, USA) based on this partial amplified fragment. For 5′-end amplification, a forward primer and a nested primer were: CYP-GSP1 (5′-CCA CCG ACT GCC GTT CAG GAA AGG-3′) and CYP-NGSP1 (5′-GGA TGT TTG ACG AAT AAA GCC GGT TG-3′), respectively. For 3′-end amplification, a forward primer and a nested primer were: CYP-GSP2 (5′-GGC TAA GCG CCC TGA CAT ACT AAA AC-3′) and CYP-NGSP2 (5′-CGT TTA CAT CCA CCG ATT TCG CTG CT-3′), respectively. The amplified PCR fragments from each reaction were purified, cloned and sequenced as described above. The resulting overlapping sequences were assembled to obtain the full-length CYP321E1 cDNA sequence.
Finally, to confirm the assembled cDNA sequence from overlapping PCR products, the entire coding region of CYP321E1 was amplified by PCR reaction with the primer pair CYP-F (5′-ATG ATT TTA GCA ATA ATA TTG TTA T-3′) and CYP-R (5′-TCA CTC TTT ATT CCT AGG TAC AAA T-3′). The following cycling parameters were used: 94 °C for 5 min followed by 35 cycles of 94 °C for 40 s, 48 °C for 30 s and 72 °C for 1 min, followed by 1 cycle at 72 °C for 10 min. PCR product was cloned and then sequenced in both directions.
Sequence analysis
The prediction of the open reading frame (ORF) was administered using ORF Finder tool at the NCBI website (http://www.ncbi.nlm.nih.gov/guide/all/#tools_). The translation of the cDNA sequence into amino acid sequence was performed using the translate tool at the ExPASy proteomic website (http://www.expasy.org/tools). The protein isoelectric point (pI) and molecular mass were predicted based on the amino acid sequence. Multiple sequence alignments of the amino sequences were made using Multiple Alignment software (http://www.phylogeny.fr/).
Expression profile of CYP321E1
The total RNA was extracted from various developmental stages and tissues of the fourth-instar larvae of P. xylostella using the same method as described above. Total RNA was collected from six developmental stages, including eggs, first-, second-, third-, fourth-larvae and pupas. Tissues used for RNA extraction were hemocytes, fat bodies, epidermis, Malpighian tubule and midgut of the fourth-instar larvae (one day after molting). Subsequently, the first-strand cDNA was synthesized from 2 μg RNA using iScript™ cDNA Synthesis Kit (Bio-rad, USA).
The pair of primers used for real-time quantitative PCR (RT–qPCR) analysis were designed and sequenced as follows: CYP-qPCR-F forward primer, 5′-TCG CCG AAA GTC TTC CAA AT-3′ and CYP-qPCR-R reverse primer, 5′-CGC TGG ATT CGT CTT TCA TC-3′ (178 bp product). RT–qPCR was performed on a CFX96 Touch™ Real-Time PCR Detection System (Bio-rad, USA) using IQ SYBR Green supermix (Bio-rad, USA). The thermal cycling profile consisted of an initial step at 95 °C for 3 min, followed by 40 cycles of 95 °C for 10 s, 45 °C for 30 s and 72 °C for 20 s. The PCR mixture (20 μl) contained 10 μl SYBR Green supermix, 0.5 μl of each primer, 1 μl of 1:20 diluted cDNA templates and sterilized water to reach the final volume. The samples were analyzed in triplicate. β-Actin (Lin et al., Reference Lin, Jin, Hu, Chen, Yin, Li, Dong, Zhang, Ren and Feng2013) was used as a reference gene to normalize the target gene expression levels among the samples.
Insecticide exposures
Although the cytochrome P450 gene CYP321E1 was one of the up-regulation detoxification genes in chlorantraniliprole resistance strain of P. xylostella, it is still necessary to evaluate the effect of different insecticides, including chlorantraniliprole and other commonly used insecticides on the expression of CYP321E1 in P. xylostella. In this study, three insecticides were used, alphamethrin, abamectin and chlorantraniliprole, each insecticide with a dose of LC15 (250, 20 and 25 μg ml−1, respectively). The leaf dip method described by Tabashnik et al. (Reference Tabashnik, Cushing and Finson1987) was adapted. In each treatment, about 20 third-instar larvae and a treated cabbage leaf were placed into the plastic Petri dish (9 cm in diameter) and kept at 25±2 °C, 65±5% RH. Each treatment was repeated three times and larvae treated with sterilized water were used as controls. After 24 h, surviving larvae were quickly frozen in liquid nitrogen. Subsequent experiments, including total RNA extraction, synthesis of the first-strand cDNA and RT–qPCR analyses were conducted as described above.
Functional analysis of CYP321E1 by RNAi
In order to obtain specific RNAi effect for the target gene, a primer pair dsCYP-F (5′-TAA TAC GAC TCA CTA TAG GG GGC TCC TAC AAT GAG TCT TA-3′) and dsCYP-R (5′-TAA TAC GAC TCA CTA TAG GG TCA AAG TCG GTT TCG TGG CA-3′) were designed for double stranded RNA (dsRNA) syntheses based on the entire coding sequence of CYP321E1. At the same time, dsGFP was synthesized as a negative control based on the sequence of pEGFP-N1 plasmid (TIANGEN, China).The primer sequences were: dsGFP-F 5′-TAA TAC GAC TCA CTA TAG GG AAG GGC GAG GAG CTG TTC ACC G-3′ and dsGFP-R 5′-TAA TAC GAC TCA CTA TAG GG CAG CAG GAC CAT GTG ATC GCG C-3′. The bold font portion of sequence was a T7 promoter sequence. Template for in vitro transcription reaction was prepared by PCR amplification using the primer pairs and the PCR products (dsCYP 580 bp and dsGFP 657 bp, respectively) and were obtained using the following cycling parameters: 95 °C for 3 min, 30 cycles of 95 °C for 40 s, 56 °C for 30 s and 72 °C for 40 s, followed by a final extension step of 72 °C for 8 min. Aliquots of 2 μl PCR products were analyzed by 1% agarose gel and the PCR product was cloned and sequenced to confirm its identity. Then the expected fragment (45 μl) was examined by 1% agarose gel and purified by Gel Extraction Kit (Omega, China). dsRNA was synthesized using T7 RiboMAX™ Express RNAi System (Promega) following the manufacturer's instructions and dissolved in nuclease-free water and examined by 1.5% agarose gel. After the concentration of dsRNA was determined by Thermo Scientific NANODROP 2000C Spectrophotometer (Thermo, USA), the final concentration of dsRNA was adjusted to 4 μg μl−1.
The fourth-instar larvae (one day after molting) were used for dsRNA injection experiments. In each treatment, ten larvae were injected with 0.5 μl dsRNA (2 μg insect−1) into the abdomen between third and fourth abdominal segments using Micro 4™ MicroSyringe Pump Controller (Sarasota, USA). The larvae injected with dsGFP were used as the negative control. Each treatment or control was repeated three times. In order to assess the expression level of each treatment, the whole body of the larvae was used for total RNA extraction. For each group, five larvae were picked out for RNAi efficiency test at two times (12 and 24 h) after injection by the RT–qPCR method.
For insecticide bioassay after RNAi, 60 larvae from the dsRNA-injected or negative control group after the 24 h injection time (the time point showed relatively high RNAi efficiency, and allowed them to feed with non-toxic leaves prior to mortality assessment) were separated into four replicates, 15 larvae per replicate and the cabbage leaf dip method described by Tabashnik et al. (Reference Tabashnik, Cushing and Finson1987) was adapted to assay the efficiency of RNAi. Each insecticide with a dose of LC50 and the procedure was conducted similarly as above described. The mortalities of the treated larvae were assessed at 48 h after insecticide treatment.
Results
cDNA cloning and analysis of CYP321E1
In the present study, a 922 bp cDNA fragment from P. xylostella was amplified and cloned by reverse-transcription PCR (RT–PCR) with specific primers. Based on this sequence, 5′- and 3′-ends RACE reactions were performed, and the full-length cDNA was obtained by overlapping the RACE cDNA fragments. The full-length cDNA sequence of CYP321E1 is 1960 bp long with an ORF of 1503 bp, which encodes a protein of 500 amino acid residues. The ORF sequence began with the first ATG codon at position 118 and ended with a TGA termination codon at 1617. The 5′-UTR and 3′-UTR were 117 and 340 bp, respectively. Based on the translated amino acid sequence of ORF, CYP321E1 has a predicted mass of 56.62 kDa and a theoretical PI value of 8.35. The heme-binding sequence motif of FGEGGRLCIG (FXXGXXXCXG) was identified at amino acid residues 436–445. The putative ‘meander’-binding sequence of ESLRLHP (EXXRXXP) and PERF (PXRF) in helix – K, the sequence motif of GQST (GXXT) in helix – I and WKLMR (WXXXR) in helix – C were also identified (fig. 1). The putative amino acid sequence of CYP321E1 has less than 50% similarity to all other amino acid sequences that already existed in GenBank, only shared optimal homology to three cDNA clones: Helicoverpa zea CYP321A1 (48% identity), Spodoptera littoralis CYP321A11 (47% identity) and Spodoptera litura CYP321B1 (44% identity) (fig. 2). Finally, the cytochrome P450 gene was named CYP321E1 (GenBank Accession No: KC 626090) by the P450 Nomenclature Committee (Dr D. Nelson). It is a novel member of CYP321 family in P. xylostella.

Fig. 1. Complete nucleotide and deduced amino acid sequence of CYP321E1 cDNA of P. xylostella. The start codon (ATG) and stop codon (TGA) are highlighted in black. The P450s signature motifs (GXXT, EXXRXXP, PXRF and WXXXR) are highlighted in gray and the heme-binding sequence motif of FXXGXXXCXG is also highlight in gray and underline in black. The sequence was deposited in the GenBank (Assession No: KC 626090).
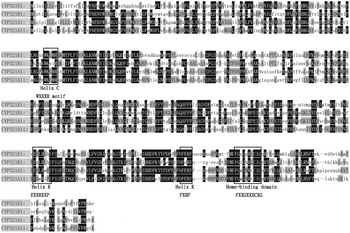
Fig. 2. Comparison of amino acid sequences of CYP321E1 (KC626090) from P. xylostella, CYP321A1 (AAM54724) from Helicoverpa zea, CYP321A11 (AFP20596) from S. littoralis and CYP321B1 (ADA68175) from Spodoptera litura, several conserved motifs of these cytochrome P450 proteins are boxed.
Expression profile of CYP321E1
The expression profiles of CYPA321E1 at different developmental stages and tissues of the fourth-instar larvae of P. xylostella were examined using RT–qPCR. β-Actin was used as an internal reference gene. The expression was detected at all developmental stages, but it was significantly higher in the fourth-instar larvae (fig. 3a). And in the fourth-instar larvae, the high expression of CYP321E1 was observed in the midgut, followed by fat bodies, and then epidermis, but its expression was relatively low in the Malpighian tubule and hemocytes (fig. 3b).
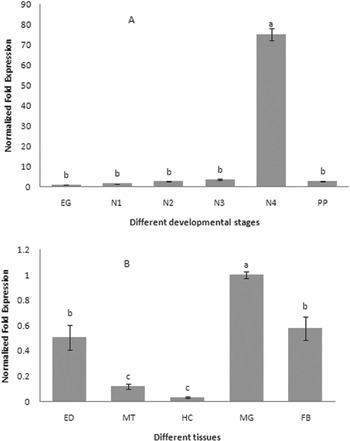
Fig. 3. Expression patterns of CYP321E1 in different tissues of the fourth-instar larvae and different developmental stages in the whole body as evaluated by RT–qPCR. (A) Stage-dependent expression pattern was examined in six different developmental stages, including eggs (EG), first- (N1), second- (N2), third- (N3), four- (N4) and pupa (PP); (B) Tissue-dependent expression pattern was examined in five different tissues, including hemocytes (HC), fat bodies (FB), epidermis (ED), Malpighian tubule (MT) and midgut (MG). β-Actin was used as an internal reference gene. Different letters on the bars indicate that the means of the gene transcript level are significantly difference among the different tissues or developmental stages based on Fisher's LSD multiple comparison test (P<0.05).
Expression response of CYP321E1 to insecticide exposures
In order to examine and confirm which insecticide could be effective at inducing the expression of CYP321E1 in P. xylostella, alphamethrin, abamectin and chlorantraniliprole were selected in this study. As the results show in fig. 4, all three insecticides increased gene expression of CYP321E1 at the dose of LC15, the maximum effect of chlorantraniliprole was observed (8.64-fold, P<0.05), followed by abamectin (2.98-fold, P<0.05). However, exposure of the insects to alphamethrin at the concentration of LC15 did not show significant effect on the expression of this gene (1.31-fold, P<0.05).
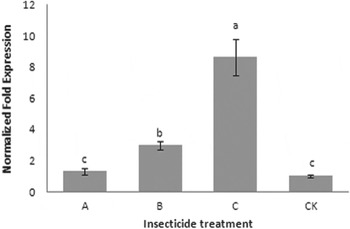
Fig. 4. Effect of alphamethrin (A), abamectin (B) and chlorantraniliprole (C) on the expression of CYP321E1 after the larvae of P. xylostella treated with each insecticide at LC15 dose: 250, 200 and 25 μg ml−1, respectively, and analyzed at 24 h by RT–qPCR. The larvae treated with sterilized water were used as controls (CK). β-Actin was used as an internal reference gene. Different letters on the bars indicate that the means of the gene transcript level are significantly difference among the different treatments based on Fisher's LSD multiple comparison test (P<0.05).
Functional analysis of CYP321E1 by RNAi
We deemed fourth-instar larvae are the best stage for RNAi not only because the relatively higher expression of CYP321E1 in this stage, but also because the size of the larvae would be suitable for administration of dsRNA by injecting. RT–qPCR analyses at 12 and 24 h after dsRNA injection of CYP321E1 showed respectively slightly and significantly reduced by about 1.33- and 5.39-fold expression levels as compared with those in the negative control (P<0.05) (fig. 5).
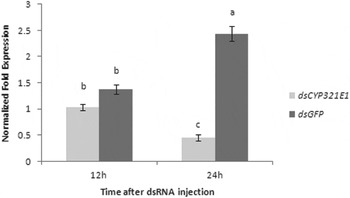
Fig. 5. Changes in the transcript levels of CYP321E1 after the fourth-instar larvae were injected with its dsRNA. Control larvae were injected with equivalent μg larvae−1 of dsGFP. Different letters on the bars indicate that the means of the gene transcript level are significantly difference among the different treatments based on Fisher's LSD multiple comparison test (P<0.05).
As the CYP321E1 expression was significantly decreased by RNAi at 24 h, we assessed RNAi effect on larvae resistance to different insecticides. The mortalities of the larvae injected with dsGFP (control) and dsCYP after treatment with chlorantraniliprole at the concentration of 50 μg ml−1 (about at the dose of LC50) were 41.7 and 70.0%, the results represent an increased mortality by approximately 1.7-fold after the repression of the CYP321E1 gene by RNAi (P<0.05). However, similar treatment with abamectin (40 μg ml−1, also about at the dose of LC50) in the larvae after RNAi for this gene did not show significant effects on the mortality of the larvae (P<0.05) (table 1).
Table 1. Toxicity of chlorantraniliprole and abamectin to the fourth-instar larvae injected with dsRNA after 24 h.

a Total number of insects used.
Asterisk after the number indicates significant difference between the mortality of the dsCYP and dsGFP treatments based on unpaired student's t-test at P<0.05.
Discussion
Previous studies indicated that up-regulation of the cytochrome P450 genes was one of the major factors leading to chlorantraniliprole resistance in P. xylostella (Lin et al., Reference Lin, Jin, Hu, Chen, Yin, Li, Dong, Zhang, Ren and Feng2013). Therefore, identification and characterization of related cytochrome P450 genes have become very attractive topics. To date, however, no cytochrome P450 gene from P. xylostella has been reported to be responsible for chlorantraniliprole resistance. Herein, we obtained the full-length cDNA sequence of a novel cytochrome P450 gene CYP321E1 by sequencing the cDNA after RACE PCR, analyzing the cDNA and deducing the amino acid sequence. Notable in the deduced amino acid sequence of this cDNA sequence is salient characteristics of cytochrome P450s that include the heme-binding decapeptide (FXXGXXXCXG), sequences EXXRXXP and PXRF in helix-K, GXXT in helix-I and WXXXR in helix-C. We also compared transcript expression of CYP321E1 between different strains, a higher expression level (74.42-fold) in the chlorantraniliprole-resistant strain by comparison to the susceptible strain was observed (data unpublished). These initial findings strengthened the possibility that CYP321E1 could be involved in chlorantraniliprole resistance in P. xylostella.
To investigate the expression profile of CYP321E1, we used RT–qPCR to analyze different developmental stages of chlorantraniliprole-resistant strain of P. xylostella. The expression of CYP321E1 was significant higher in the fourth-instar larvae than the larvae of other developmental stages. The different tissues of the fourth-instar larvae were further analyzed, CYP321E1 was found to be most abundant in the midgut. Chlorantraniliprole is both a stomach and contact toxicant, suggesting that higher expressions of CYP321E1 in midgut, fat bodies and epidermis may correlate well with how these tissues encounter insecticides. In insects, midgut and fat bodies are generally thought to be the major detoxification organs (Hodgson, Reference Hodgson, Kerkut and Gilbert1985; Snyder et al., Reference Snyder, Stevens, Andersen and Feyereisen1995; Bautista et al., Reference Bautista, Miyata, Miura and Tanaka2009). Our findings of tissue-dependent expression patterns are consistent with this notion for CYP321E1. However, since the epidermis also exhibited relatively high CYP321E1 expression, it is possible that initial detoxification by CYP321E1 of chlorantraniliprole absorbed through contact with residues on cruciferous vegetable surfaces, also occurs here. These results may imply an adaptive ability of P. xylostella larvae to metabolize chlorantraniliprole upon exposure and the fourth-instar larvae could be important in insecticide resistance.
It is possible that insecticide resistance is due to interactions of a particular insecticide with the regulatory mechanism of induction (Liu & Scott Reference Liu and Scott1996; Le Goff et al., Reference Le Goff, Hilliou, Siegfried, Boundy, Wajnberg, Sofer, Audant, ffrench-Constant and Feyereisen2006). Recognizing this possibility, we also attempted to examine the inducibility of CYP321E1 gene by alphamethrin, abamectin and chlorantraniliprole in P. xylostella. The low induction concentrations of insecticides and exposure duration in our study were determined according to the report of Guo et al. (Reference Guo, Zhang, Yu, Zhu, Guo and Ma2012), for that insecticides could play important roles in intoxication rather than induction under high concentration. The induction data show that CYP321E1 responded differently to the exposures of insect larvae to three different insecticides. Chlorantraniliprole significantly induced the expression of CYP321E1 at 24 h, abamectin also appeared to induce the expression of CYP321E1, but the induction effect was significantly less than that in chlorantraniliprole treated insects. Alphamethrin at present induction concentration and duration did not affect the expression of CYP321E1. So we inferred that the up regulation of CYP321E1 mRNA in P. xylostella larvae was correlated with development of chlorantraniliprole resistance because significant increases in the LC15 values were detected. These findings are also in agreement with our precious studies on transcriptome analysis of chlorantraniliprole resistance development in P. xylostella (Lin et al., Reference Lin, Jin, Hu, Chen, Yin, Li, Dong, Zhang, Ren and Feng2013).
However, all the above-mentioned findings cannot pinpoint whether or not CYP321E1 is actually involved in insecticide resistance. However, it has been suggested that the major enzymes involved in insecticide detoxification could be identified by a means of induction profiling of insect detoxification enzymes (Poupardin et al., Reference Poupardin, Reynaud, Strode, Ranson, Vontas and David2008). In order to further confirm whether or not the chlorantraniliprole inducible gene CYP321E1 is involved in the chlorantraniliprole detoxification, we carried out RNAi to silence the target gene by injecting the sequence-specific dsRNA to the fourth-instar larvae of P. xylostella followed by insecticides bioassay. A noticeable reduction of the CYP321E1 expression in the larvae after RNAi 24 h was observed and analysis of knockdown effect of the gene on resistance, based on the bioassay, indicated that CYP321E1 is responsible for up to 70.0% mortality, which corresponds to 1.7-fold reduction in resistance to for dsRNA-injected larvae. However, from the present data, mortality of abamectin bioassay was not significantly different between dsRNA-injected and control larvae, indicating that CYP321E1 is not likely involved in abamectin detoxification in P. xylostella. Of course, the data on resistance of CYP321E1 RNAi also suggested that chlorantraniliprole resistance is not due to CYP321E1 alone, mainly because resistance was not completely abolished. The remaining resistance in P. xylostella could be due to other mechanisms such as target-site mutation in the C-terminal membrane-spanning domain of the RyR (Troczka et al., Reference Troczka, Zimmer, Elias, Schorn, Bass, Davies, Field, Williamson, Slater and Nauen2012).
In summary, we identified and characterized a novel cytochrome P450 gene, which belongs to a cytochrome P450 gene family CYP321, from an important agriculture pest P. xylostella. The findings indicate that overexpressed CYP321E1 is involved in increased chlorantraniliprole detoxification partly leading to resistance. So it is important to further study the difference in the metabolism ability of CYP321E1 to chlorantraniliprole between the susceptible strain and the resistance strain, and investigate the enzyme properties of this gene in both strains. These studies will influence the practice of chlorantraniliprole resistance management strategies for P. xylostella.
Acknowledgements
This research was supported by Guangdong Natural Science Foundation (Grant numbers S2013010012529 and S2013040015379), Special Fund for Agro-scientific Research in the Public Interest (Grant No: 201103021), Guangzhou Municipal Science and Technology planning Project (Grant No: 2014J4100197)and President Foundation of Guangdong Academy of Agricultural Sciences (Grant No: 201206). The authors give special thanks to Dr D. Nelson for help in the nomenclature of the cytochrome P450 gene.








