Fish have the ability to elongate and/or desaturate fatty acid precursors to synthesise long-chain (LC) PUFA(Reference Sargent, Tocher, Bell, Halver and Hardy1), but the efficiency of this metabolic pathway varies among fish species. In contrast to marine finfish species where the ability of synthesising LC-PUFA from precursors is limited and controversial(Reference Monroig, Zheng and Morais2, Reference Tocher and Ghioni3), freshwater fish seem to be able to maintain a certain degree of Δ6-, Δ5-desaturase and elongase activities to produce arachidonic acid, EPA and DHA if their precursors linoleic and α-linolenic acids (ALA) are present in their diet. Fatty acid desaturase 2 (FADS2), also called Δ6-desaturase, is the rate-limiting enzyme in the production of EPA and DHA from ALA(Reference Cook and Vance4) even if two other enzymes (FADS1/Δ5-desaturase and elongase) are involved in this biosynthetic pathway. FADS2 is a membrane-bound protein with amino-terminal cytochrome b5 domains carrying haem-binding motifs, two-membrane-spanning domains and three His-box motifs(Reference Cho, Nakamura and Clarke5). FADS2 converts ALA (18 : 3n-3) into stearidonic acid (18 : 4n-3) and linoleic acid (18 : 2n-6) into γ-linolenic acid (18 : 3n-6), which are the first step in the pathway of n-3 and n-6 LC-PUFA synthesis, respectively.
Δ6- and Δ5-Desaturases are of major interest in the field of fish nutrition, especially with regard to the possibility of dietary linoleic and ALA to fulfil the requirement of teleosts for the essential LC-PUFA of the n-3 and n-6 series. Until recently, LC-PUFA requirements have been satisfied by lipids from fishery-derived products, but such LC-PUFA sources have reached their limit of sustainability(Reference Pike and Barlow6, Reference Shepherd, Pike and Barlow7). The ability to convert linoleic and α-linolenic fatty acids, abundant in alternative lipid sources such as vegetable oils, into LC-PUFA is well established in some species, such as rainbow trout(Reference Owen, Ardon and Middleton8–Reference Seiliez, Panserat and Kaushik12), but has been poorly investigated in other freshwater species, including European grayling(Reference Tulli, Cardinaletti and Messina13). Individual variability in the capacity of fish to retain or synthesise n-3 LC-PUFA when fed vegetable oil diets has been shown in Atlantic salmon(Reference Schlechtriem, Bron and Tocher14). Moreover, Leaver et al. (Reference Leaver, Taggart and Villeneuve15) demonstrated that flesh n-3 LC-PUFA composition is a highly heritable trait (h 2= 0·77) in salmon. However, in their study, Morais et al. (Reference Morais, Taggart and Guy16) showed that the hepatic expression of Δ6-desaturase was not responsible for the variations observed in the levels of n-3 LC-PUFA in four families of Atlantic salmon. Therefore, these authors have suggested that the observed variability might originate from the presence of different alleles of fatty acyl desaturases encoding proteins with altered biological activity or specificity.
Recent findings have outlined that genetic variation in FADS1 and FADS2 gene clusters is associated with altered desaturase activity in human subjects(Reference Merino, Johnston and Clarke17). Moreover, FADS2 polymorphism has been shown to modulate the activity of Δ6-desaturase in human subjects(Reference Merino, Ma and Mutch18), Japanese quails(Reference Khang, Jennen and Tholen19) and pigs(Reference Renaville, Prandi and Fan20); however, the association with desaturase activity has not yet been examined in fish. Therefore, FADS2 is a candidate gene for increasing EPA and DHA synthesis from ALA in fish.
Thus, the present study aimed to study the Δ6-desaturase polymorphism and its impact on the muscle fatty acid profile in European grayling (Thymallus thymallus L., 1758), chosen as a cold freshwater fish model. This fish species has a natural distribution covering vast parts of Europe, from France and England eastwards to the Ural Mountains and from Scandinavia southwards to the Balkan region and Northern Italy(Reference Northcote21). Actually, there is a growing interest for this species in fisheries management and conservation plans (European Dir. Habitat 92/43/CEE) and, recently, also in aquaculture.
Materials and methods
Animal and rearing conditions
For the present study, 100 specimens of European grayling (initial body weight 12·1 (SD 0·1) g) were obtained from the hatchery Ribiška Druzina (Tolmin, Slovenija). Fish originated from a single stock were obtained by mating one 4-year-old female and two males. They were stocked in a 500-litre fibreglass tank in the experimental flow-through facilities of the Department of Food Science at the University of Udine (Italy) supplied with well water (water temperature 13·7 ± 0·22°C; dissolved oxygen 8·17 (sd 0·8) mg/l; pH 7·49 ± 0·21; total ammonia nitrogen 0·08 (sd 0·01) mg/l; photoperiod 12 h light–12 h dark). After stocking, fish were hand-fed a commercial diet for trout (feed formula: 42 % crude protein, 25 % crude lipid on dry feed, Dibaq ATX 5HMDI; Dibaq Italia) in two daily meals (09.00 and 16.00 hours) until the first feed item was refused, over 64 d. At the end of the trial, fish were weighed (final weight 31·8 (SD 2·5) g) under moderate anaesthesia (5 mg benzocaine/l alcohol solution) and twenty-seven specimens were killed with excess of the anaesthetic (30 mg benzocaine/l alcohol solution) to obtain the fillet. Handling of the fish was done according to the guidelines of the European Union Directive (2010/63/EU) on the protection of animals used for scientific purposes.
Molecular genetic marker evaluation
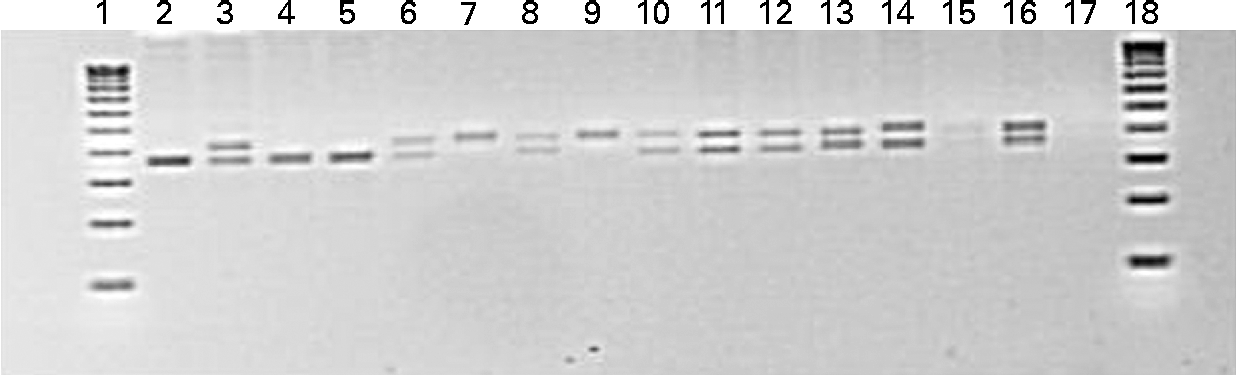
DNA was extracted from muscle samples (twenty-seven animals) using a standard DNA extraction method (GenElute Mammalian Genomic DNA Miniprep Kit; Sigma-Aldrich). A primer set (forward: AAAGAAGTTGAAGTACATGCCCTA; reverse: AGTTCCGTTGTGAAAACATGG) was designed in order to amplify a fragment of the FADS2 gene based on the Salmo salar sequence (GenBank accession no. AY736067.2). The forward primer was designed in S. salar exon 7 and the reverse one in exon 8. PCR conditions consisted of an initial step at 94°C for 3 min followed by forty cycles of 94°C for 30 s, annealing temperature (60°C) for 30 s and 72°C for 30 s. The final extension step consisted of 5 min at 72°C. PCR products were separated on a 2·5 % agarose gel in 1 × TBE buffer by electrophoresis. Separated fragments on agarose gels were sized by referencing with a 100 bp DNA ladder (Fig. 1). The bands were cut and DNA extracted with a gel extraction kit (Zymoclean Gel DNA Recovery Kit; Zymo Research). PCR products were sequenced from the gel extract using an ABI automated DNA sequencer (Applied Biosystems).

Fig. 1 PCR products of fatty acid desaturase 2. First and last lanes: molecular weight (100 bp/band); second, fourth and fifth lanes: homozygous for the deletion; seventh and ninth lanes: homozygous for the insertion; other lanes: heterozygous.
Sequence comparison
The sequence of European grayling (T. thymallus) was compared with the sequence of Atlantic salmon (S. salar, GenBank accession no. AY736067.2), cod (Gadus morhua, Ensembl ENSGMOG00000017746) and zebrafish (Danio rerio, Ensembl ENSDARG00000019532) using BLAST (bl2seq) from NCBI.
Lipid analysis
Muscle lipids were extracted with a chloroform–methanol (2:1, v/v) solution containing 0·01 % butylated hydroxytoluene according to Folch et al. (Reference Folch, Lees and Sloane Stanley22).
A 5 % solution of HCl in methanol was used to prepare fatty acid methyl esters according to Christie(Reference Christie23). Fatty acid methyl esters were separated and quantified by high-resolution GC (TRACE GC2000; Fisher Scientific SAS) using a 30 m × 0·32 mm (inner diameter) Omegawax™ Capillary GC Column (Supelco) with flame ionisation detection at 250°C. The carrier gas (H2) flow rate was 1 ml/min, and samples (1 μl) were directly injected in a split ratio of 1:30. The oven temperature was programmed from 180°C for 6 min, then at 3°C/min until 225°C and held for 10 min. The chromatograms were acquired and processed using ChromQuest integration software (ThermoQuest Italia Limited) and individual methyl esters were identified by comparison with known standards obtained from Supelco, Inc. Repeated injections of the standard solution were carried out to test the analytical precision. All solvents and reagents were of analytical grade and were purchased from Sigma-Aldrich, Inc. Data on individual fatty acid composition are expressed as a percentage of total identified fatty acids.
Data analysis
Insertion/deletion associations with fatty acid proportions were analysed using the general linear mixed model procedure of SPSS version 17 (SPSS, Inc.) with a model that included insertion/deletion as a fixed effect. Significant differences were declared when the marker genotype effect was a significant source of variation in ANOVA, and the P value for the difference between the least-squares means for each marker genotype was less than 0·05. If appropriate, the post hoc least significant difference test was applied to compare the means.
Results
Sequencing revealed an insertion/deletion polymorphism in the seventh intron of FADS2. The insertion consists of eleven repeats of CTGT (44 bp); indeed, in this intronic region, the motif CTGT is repeated either forty times (insertion allele) or twenty-nine times (deletion allele). The allelic frequency was 39 % for the insertion (obtained by adding half of the heterozygous frequency to the homozygous frequency) and 61 % for the deletion.
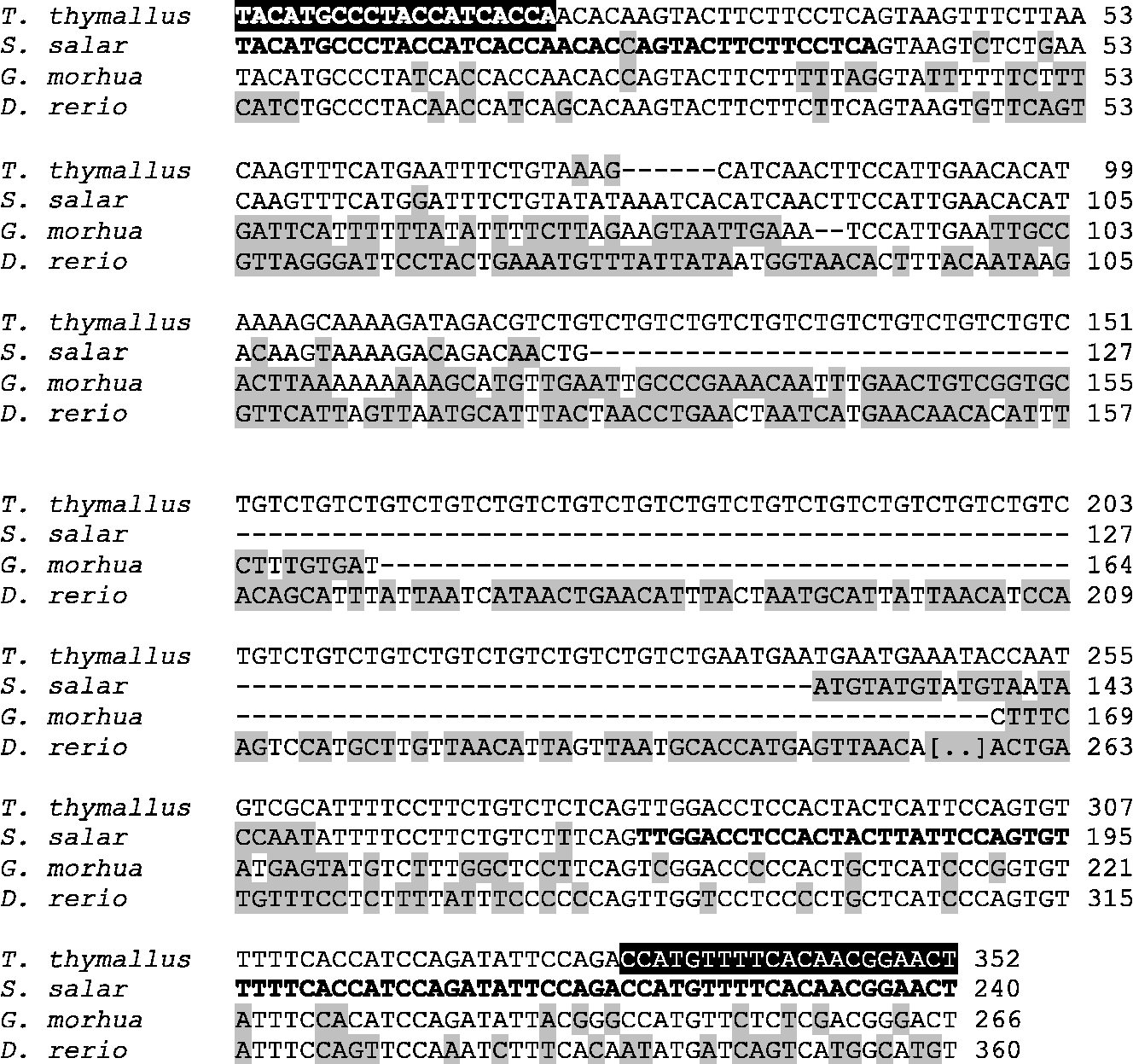
The sequence for the deletion variant of the amplified fragment of FADS2 of European grayling is reported in Fig. 2 along with Atlantic salmon (S. salar), cod (G. morhua) and zebrafish (D. rerio) sequences. With respect to the exons, the intronic region was more dissimilar between salmon and grayling and the repeated motif was not present in S. salar. The grayling sequence of the exonic region was quite similar to that of D. rerio and G. morhua, while the intronic region was completely dissimilar.

Fig. 2 Comparison of the nucleotide sequence of fatty acid desaturase 2 (FADS2) from Thymallus thymallus (deletion variant) with that of FADS2 from Salmo salar, Gadus morhua and Danio rerio. Sequences were aligned using Blast (bl2seq from the NCBI). White letters, black background: primers used; bold: salmon exons; grey background: sequence differences from T. thymallus.
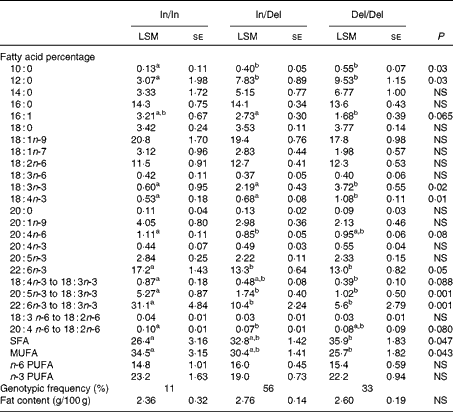
The total lipid content of the fish fillets and the fatty acid composition associated with the observed polymorphism are presented in Table 1.
Table 1 Genotypic least-squares means for fatty acid desaturase 2 on total lipid content (g/100 g muscle) and fatty acid concentrations (% total fatty acids) (Least-squares mean values with their standard errors)

In, insertion; Del, deletion.
a,bMean values within a row with unlike superscript letters were significantly different (P< 0·05).
The FADS2 polymorphism is associated with clear differences in the incidence of specific fatty acid ratio along the desaturation pathway that almost reached significance in the case of 18 : 4n-3 to 18 : 3n-3 (P= 0·088), but resulted highly significant in the case of 20 : 5n-3 to 18 : 3n-3 (P= 0·001) and 22 : 6n-3 to 18 : 3n-3 (P= 0·001). Such a clear effect of the polymorphism was not observed in the ratio of n-6 fatty acids, although the activity of Δ6-desaturase seemed to be lower in the Insertion/Insertion fish. The mutation is also associated with significant differences in palmitoleic acid, ALA, stearidonic acid and DHA percentages as well as SFA and MUFA.
Discussion
In the present study, we revealed the presence of a polymorphism in the FADS2 gene of T. thymallus. The repeated motif where we observed the insertion/deletion polymorphism was not present in S. salar nor in G. morhua or in D. rerio sequences, suggesting that if these species present polymorphisms in the FADS2 gene, they will be different.
The effect of the FADS2 polymorphism on Δ6-desaturase activity was first observed in human studies(Reference Martinelli, Girelli and Malerba24) where an association with the EPA:ALA ratio was also outlined. Moreover, in European adolescents, one minor allele of the FADS1–FADS2 gene cluster was associated with higher Δ6-desaturase activity levels(Reference Bokor, Dumont and Spinneker25). More recently, FADS2 polymorphisms have also been observed in other mammals, such as pigs(Reference Renaville, Prandi and Fan20), where the authors have also shown a modulation of Δ6-desaturase activity. The insertion/deletion polymorphism in the seventh intron of the FADS2 gene of T. thymallus observed in the present study affected DHA and ALA proportions in fillet lipids, which is in agreement with studies on human subjects showing that SNP in the FADS1–FADS2 gene cluster reduces EPA concentrations and increases ALA concentrations in plasma phospholipids(Reference Merino, Johnston and Clarke17, Reference Lemaitre, Tanaka and Tang26), but also in breast milk(Reference Xie and Innis27) and adipose tissue(Reference Baylin, Ruiz-Narvaez and Kraft28). Several other studies in human subjects have shown the associations between mutations in the FADS1–FADS2 gene cluster and fatty acid composition (reviewed in Merino et al. (Reference Merino, Ma and Mutch18)). Consistently, Khang et al. (Reference Khang, Jennen and Tholen19) observed significant associations between the FADS2 polymorphism and the DHA content in the egg yolk of Japanese quail.
Also in the present study, the ratio of products:precursors was used to assess the activity of Δ6-desaturase, as already proposed by Merino et al. (Reference Merino, Johnston and Clarke17). The insertion of the repeated motif resulted in an increase of Δ6-desaturase activity in fatty acid biosynthetic pathways for n-3 PUFA. However, the polymorphism does not seem to affect the activity of the enzyme on n-6 PUFA. This might result from a different activity of the enzyme on the different precursors (n-3 or n-6), as it has been observed in dogs(Reference Dunbar and Bauer29) and Murray cod(Reference Francis, Peters and Turchini30). Moreover, Thanuthong et al. (Reference Thanuthong, Francis and Senadheera31) showed that, in the trout, the Δ6-desaturase has a 3·2-fold higher affinity towards 18 : 3n-3 over 18 : 2n-6. Despite a relatively limited number of animals analysed in the present study, these preliminary results suggest that the genetic selection of European grayling for this marker might improve its ability to utilise dietary LC-PUFA precursors, as FADS2 is the rate-limiting enzyme in the production of EPA and DHA from ALA.
As nutritionists have recommended the consumption of fish for its beneficial properties due to the high content in LC-PUFA, namely EPA and DHA, which have a variety of important physiological functions(Reference Ruxton, Reed and Simpson32–Reference Turchini, Nichols and Barrow36), a similar approach applied to other finfish species of aquaculture interest may, by using marker-assisted selection, allow an improvement in the utilisation of dietary vegetable oils and meals, rich in n-3 precursors, and also provide human consumers a healthier fish product.
Acknowledgements
The present study was supported by Ente Tutela Pesca – Regione Friuli Venezia Giulia (Italy). The authors are thankful to Aysum Adakli, Enrico Daniso and Massimo Lanzoni for their precious technical assistance. B. R. and M. M. developed the initial idea, designed the study and were in charge of the genetic determinations; F. T. was in charge of the lipid analysis; M. B. was responsible for the fish trial; E. T. supervised the study at all steps. All the authors critically revised the drafted manuscript. None of the authors has any financial or personal conflicts of interest.