Research carried out in the past few years has highlighted the importance of the gut microbiota to intestinal health and host health in general( Reference Guarner 1 , Reference Sekirov, Russell and Antunes 2 ). The gut microbiota plays a crucial role in the development and maintenance of the immune system( Reference Kau, Ahern and Griffin 3 ), colonisation resistance to environmental bacteria, such as pathogens( Reference Stecher and Hardt 4 ), extraction of energy from indigestible food components and production of metabolites that influence gut health( Reference Louis, McCrae and Charrier 5 ). SCFA have been widely investigated due to their ability to influence several aspects of host health. Many members of the gut microbiota produce acetate and propionate. Acetate is either reused by other bacteria for butyrate formation or absorbed by the host and can be used as an energy source, while propionate is involved in gluconeogenesis( Reference Hoyles, Wallace and Timmis 6 , Reference Scott, Gratz and Sheridan 7 ). The most investigated end metabolite of the gut microbiota is probably butyrate due to its importance to mucosal health. Butyrate is mainly produced by the members of Clostridium clusters IV and XIVa, such as Faecalibacterium prausnitzii, Eubacterium hallii and Roseburia spp., ( Reference Louis and Flint 8 , Reference Pryde, Duncan and Hold 9 ) and is absorbed by the mucosa where it can act as an energy source for colonocytes( Reference Roediger 10 ). Moreover, butyrate has anti-inflammatory effects, is able to promote apoptosis in cancer cells and has been shown to inhibit virulence in enteropathogens( Reference Hamer, Jonkers and Venema 11 , Reference Gantois, Ducatelle and Pasmans 12 ).
Gut microbiota composition and metabolic activity not only affect the host but are also strongly modified by host and environmental factors. Diet composition, such as fibre content, has been shown to have a strong impact on the gut microbiota( Reference Flint, Duncan and Scott 13 , Reference De Vuyst and Leroy 14 ). Recent studies have elucidated the differences in gut microbial consortium in populations living in developed and developing countries possibly due to the effects of diet or high prevalence of malnutrition in developing countries, which can strongly influence the gut microbiota( Reference Kau, Ahern and Griffin 3 , Reference Gupta, Mohammed and Ghosh 15 – Reference De Filippo, Cavalieri and Di Paola 20 ). One of the most common nutritional deficiencies is Fe deficiency, affecting more than two billion people worldwide, especially in developing countries( Reference Zimmermann and Hurrell 21 , 22 ). Fe deficiency affects cognitive and motor development in children and increases the maternal/perinatal mortality rate( Reference Lynch 23 ). It also has an impact on gut microbiota composition and metabolic activity. In a recent rat study, we have shown a decrease in the abundance of Roseburia spp./Eubacterium rectale and Bacteroides spp. along with a large decrease in the concentrations of caecal butyrate and propionate in highly Fe-deficient young rats( Reference Dostal, Chassard and Hilty 24 ). Earlier studies in mice have also demonstrated that low gut luminal Fe concentrations have an impact on gut microbiota composition leading to a decrease in the abundance of Desulfovibrio spp. and an increase in that of Turicibacter spp.( Reference Werner, Wagner and Martinez 25 ) while elevating the counts of lactobacilli( Reference Tompkins, O'Dell and Bryson 26 ). Using an in vitro colonic fermentation model mimicking the proximal colon of a child, we have also confirmed strong dysbiosis of the gut microbiota under very-low-Fe conditions( Reference Dostal, Fehlbaum and Chassard 27 ).
Usual strategies for Fe-deficiency anaemia correction are Fe supplementation and Fe fortification of foods; FeSO4 is often used, which is highly bioavailable( Reference Hilty, Arnold and Hilbe 28 ). However, absorption of Fe is usually low (5–20 %) and takes place mainly in the duodenum, while the main fraction of Fe reaches the colon, where it might affect the gut microbiota( Reference Zimmermann and Hurrell 21 ). Indeed, gut microbiota composition in Fe-deficient rats supplemented with Fe has been found to be partially recovered and gut microbiota metabolic activity to be strongly promoted( Reference Dostal, Chassard and Hilty 24 ). Animal studies in rats and pigs have also reported dysbiosis of the gut microbiota due to Fe supplementation( Reference Tompkins, O'Dell and Bryson 26 , Reference Lee, Shinde and Choi 29 ), recently confirmed in human studies. School children in Côte d'Ivoire have been found to have higher numbers of Enterobacteriaceae and decreased numbers of lactobacilli in faeces after Fe supplementation for 6 months and increased concentrations of calprotectin, a marker for gut inflammation ( Reference Zimmermann, Chassard and Rohner 30 ). Infants given an Fe-supplemented diet for 3 months have been found to have higher relative abundances of Bacteroides spp. and lower abundances of lactobacilli( Reference Krebs, Sherlock and Westcott 31 ).
An unanswered question raised by these studies is whether dietary Fe alone is a modulation factor for the gut microbiota and intestinal inflammation or whether Fe status and susceptibility to gut inflammation could also play a role in the observed changes in gut microbiota composition. Both changes in host Fe homeostasis and gut inflammation can alter the gut microbiota composition( Reference Buhnik-Rosenblau, Moshe-Belizowski and Danin-Poleg 32 – Reference Winter, Lopez and Baumler 34 ). Our previous study using Sprague–Dawley rats with a rodent microbiota has investigated the changes in gut microbiota composition due to Fe supplementation in a highly Fe-deficient host( Reference Dostal, Chassard and Hilty 24 ). As the rodent microbiota differs from the human microbiota( Reference Dethlefsen, McFall-Ngai and Relman 35 – Reference Krych, Hansen and Hansen 37 ), in the present study, we used gnotobiotic rats associated with child microbiota, which represent an excellent model to study host–microbiota interactions in a well-controlled environment without confounding factors such as variations in diet or host genetic background( Reference Hanske, Engst and Loh 38 – Reference Crouzet, Gaultier and Del'homme 40 ). A child was selected as the faecal donor due to the high prevalence of Fe deficiency and need for Fe supplementation in this age group( 22 , Reference Iannotti, Tielsch and Black 41 ). The human microbiota-associated rats were fed diets differing in Fe concentration and source according to the standard Fe depletion–repletion study used to assess Fe bioavailability( Reference Forbes, Arnaud and Chichester 42 ). Changes in faecal microbiota composition were analysed during the study period, and caecal microbiota composition and metabolic activity, as well as gut inflammation, were assessed after killing the rats at the end of the study.
Materials and methods
Rats and diets
A total of forty germ-free female Fischer 344 rats were bred at the INRA facilities of Clermont-Ferrand-Theix, France. They were kept in sterile isolators with positive pressure over the entire trial period as described previously( Reference Crouzet, Gaultier and Del'homme 40 , Reference de Sablet, Chassard and Bernalier-Donadille 43 ). Rats were fed an irradiated standard diet for germ-free rodents (SAFE) and given free access to sterilised water. Rats were maintained pairwise in standard Macrolon cages under a 12 h light–12 h dark cycle under constant temperature and humidity. At 5 weeks of age, rats were inoculated with a human faecal microbiota slurry obtained from a healthy volunteer (age 6 years) not treated with antibiotics 3 months before faecal sample collection. The faecal sample was processed within 6 h of defecation and maintained under anaerobic conditions. After 1000-fold dilution with an anaerobic mineral solution, 1 ml of faecal slurry was orally inoculated into germ-free rats. The microbiota was allowed to establish for 3 weeks while the rats were being fed a control diet with normal Fe concentrations equivalent to a standardised American Institute of Nutrition (AIN)-93G purified diet( Reference Reeves, Nielsen and Fahey 44 ) before starting the different feeding regimens.
All diets were produced by Dyets Inc. and based on a standard AIN-93G diet differing only in Fe concentration (Table S1, available online). The study set-up (Fig. 1) was designed according to the classical Fe depletion–repletion assay of Forbes et al. ( Reference Forbes, Arnaud and Chichester 42 ) and comprised five different groups of rats. A ‘control’ group of rats (n 8) was fed a regular AIN-93G diet containing a mean of 37·6 (sem 0·9) mg Fe/kg diet from ferric citrate over the entire trial period of 16 weeks, while a ‘Fe-deficient’ group of rats (n 8) was fed an Fe-deficient diet containing 2·9 (sem 0·2) mg Fe/kg diet. A further two groups of rats, ‘35 ppm Fe’ group (n 8) and ‘70 ppm Fe’ group (n 8), were first fed a Fe-deficient diet for 12 weeks and were then supplemented with either a 35 ppm Fe diet containing 32·2 (sem 1·0) mg Fe/kg diet from FeSO4 or a 70 ppm Fe diet containing 66·1 (sem 7·8) mg Fe/kg diet from FeSO4 and ferric citrate to mimic Fe supplementation with two different Fe sources and concentrations for 4 weeks. A fifth group of rats, ‘Fe-excess’ group (n 8), was first fed the control diet and was then supplemented with the 70 ppm Fe diet to mimic the impact of Fe supplementation on the gut microbiota not previously affected by a low-Fe diet and to investigate the effects of excess Fe on the gut microbiota. Once a week, body weight of each rat was measured and diet intake was assessed cage-wise (two rats). Fe concentration in the diets was assessed by atomic absorption spectrometry (SpectrAA-240K with GTA-120 Graphite Tube Atomizer, Varion Techtron) shortly before use. All procedures were carried out according to the European Directives on the protection of animals used for scientific purposes, 2010/63/EU, and the laboratory procedures were approved by the local ethics committee (CEMEAA 02).

Fig. 1 Study set-up with different iron feeding regimens according to a classical iron depletion–repletion study design. Germ-free Fischer 344 rats (n 40) were divided into five groups and inoculated with the same microbiota from a human volunteer. After 3 weeks of initial colonisation for gut microbiota establishment, diets differing only in iron concentration were fed to rats as outlined in the figure.
Sample collection
Faecal samples and blood samples of all rats were collected at baseline (week 3), after the first feeding period at midpoint (week 15) and at endpoint (week 19). Faecal samples were frozen at − 80°C until gut microbiota composition analysis by quantitative PCR (qPCR). Fe concentration in the faecal samples of the 35 ppm Fe-supplemented group, Fe-deficient group and control group at endpoint (week 19) was assessed by atomic absorption spectrometry (SpectrAA-240K with GTA-120 Graphite Tube Atomizer, Varion Techtron). Blood samples were collected by the tail vein clip method( Reference Abatan, Welch and Nemzek 45 ) and analysed immediately for Hb concentrations using a Hemocue 201 instrument (HemoCue). After killing the rats by CO2 inhalation, blood was collected again by cardiac puncture and centrifuged immediately, and serum was kept at − 80°C until ferritin measurements. Serum ferritin concentrations were assessed in duplicate using a rat ferritin ELISA kit according to the manufacturer's instructions (Immunology Consultants Laboratory, Inc.). Sections of the ileum, caecum and colon were removed, rinsed in PBS and immersed in 4 % paraformaldehyde, and stored at 4°C until histological analyses. Caecal contents were also collected and frozen immediately at − 80°C for caecal microbiota metabolic activity and composition analyses.
Histological analyses of ileal, caecal and colonic tissue samples
All histological analyses were carried out by the Histology Analysis Platform of the Nantes Atlantic National College of Veterinary Medicine, Food Science and Engineering (Oniris, Nantes, France). After sectioning, ileal, caecal and colonic tissue samples were stained with haematoxylin–eosin–safranin and analysed microscopically using a Nikon Eclipse 5DI microscope connected to a Nikon DS-42-RI1 camera (200 × or 400 × magnification) (Nikon Instruments). Tissue samples were analysed for infiltration of immune cells, damage in crypt architecture, hyperaemia and mucosal erosion and were given a histological colitis score according to the severity of these parameters( Reference Engel, Kellermann and Rau 46 ). The veterinary pathologist examining the tissue samples was blinded to treatment.
Faecal and caecal microbiota composition analysis using quantitative PCR
Total genomic DNA from faecal samples and caecal content samples was extracted using the FastDNA SPIN Kit for Soil (MP Biomedicals). For the enumeration of total 16S rRNA gene copies and nine bacterial groups prevalent in the gut microbiota, namely Firmicutes, Bacteroides spp., Clostridium cluster IV, F. prausnitzii, E. hallii, Enterobacteriaceae, Lactobacillus/Leuconostoc/Pediococcus spp., Roseburia spp./E. rectale, and sulphate-reducing bacteria (SRB), primers targeting the 16S rRNA gene or a functional gene were used for qPCR carried out with an ABI PRISM 7500-PCR sequence detection system using 2 × SYBR Green PCR Master Mix (Life Technologies), as described previously( Reference Dostal, Fehlbaum and Chassard 27 , Reference Zihler, Gagnon and Chassard 47 ). In short, PCR comprising 0·2 μm of each primer in 25 μl volume were carried out in duplicate for each sample. In every run, a standard curve with serially diluted 16S rRNA gene or functional gene concentrations of a representative strain for each bacterial target group was included. SRB were enumerated with the same protocol using the primers dsrA_F336 (5′-CTG CGA ATA TGC CTG CTA CA-3′) and dsrA_R533 (5′-TGG TCG ARC TTG ATG TCG TC-3′) targeting the dissimilatory sulphite reductase subunit A( Reference Pereyra, Hiibel and Prieto Riquelme 48 ), and E. hallii 16S rRNA gene copies were evaluated using the primers EhalF (5′-GCGTAGGTGGCAGTGCAA-3′) and EhalR (5′-GCACCGRAGCCTATACGG-3′)( Reference Ramirez-Farias, Slezak and Fuller 49 ).
Pyrosequencing analysis
Pyrosequencing analysis was carried out using caecal samples of the control group, Fe-deficient group and 35 ppm Fe-supplemented group for three rat pairs in each group. Caecal content samples of the two rats housed in the same cage were pooled, and DNA was extracted using the FastDNA SPIN Kit for Soil (MP Biomedicals), resulting in three genomic DNA samples per analysed group of rats (total of nine genomic DNA samples). Pyrosequencing analysis was carried out by DNAVision (Charleroi, Belgium) using a 454 Life Sciences Genome Sequencer FLX instrument (Roche Applied Science), following previously described procedures( Reference Dostal, Fehlbaum and Chassard 27 ). Resulting reads from the hypervariable gene region V5–V6 of the 16S rRNA gene were assigned to samples according to their multiplex identifier tag, checked for the presence of primer sequences and fragment length >200 bp. All reads not fulfilling these criteria were discarded, resulting in an average number of sequences of 5345 (sd 1929) per sample used in taxonomic assignment. Sequences were assigned on family and genus level using the Mothur( Reference Schloss, Westcott and Ryabin 50 ) and Greengenes 16S reference database( Reference DeSantis, Hugenholtz and Larsen 51 ). Moreover, sequences that did not align where 95 % of the other sequences aligned were discarded. Chimera candidates were identified using the UCHIME implementation in Mothur( Reference Edgar, Haas and Clemente 52 ). Relative abundances of unassigned reads or reads assigned on family or genus level were calculated from the total number of reads matching the quality control criteria.
Metabolite analysis
Caecal content samples were centrifuged (10 000 g ) and supernatants were diluted with MilliQ water before analysis by HPLC as described previously( Reference Cleusix, Lacroix and Vollenweider 53 ). Fermentative metabolites (lactate, formate, acetate, propionate, butyrate, isovalerate, isobutyrate and valerate) and SCFA (acetate, propionate and butyrate) in each sample were analysed in duplicate.
Statistical analysis
Statistical analysis was carried out using JMP 8.0 and SPSS 18.0 (IBM SPSS, Inc.). All data were tested for normal distribution using the Shapiro–Wilk test and are expressed as means with their standard errors. Hb, ferritin, weight gain and gut microbiota concentrations for each metabolite and caecal qPCR data for each bacterial target were compared between the groups of rats using one-way ANOVA with a post hoc Bonferroni test. Diet consumption was compared using non-parametric Kruskal–Wallis test. qPCR data on faecal microbiota composition were analysed over time for every bacterial target within each rat group using one-way repeated-measures ANOVA with Greenhouse–Geisser correction and post hoc Bonferroni test. qPCR data were log10-transformed before statistical analysis. Histological colitis scores and pyrosequencing data were compared pairwise using the non-parametric Mann–Whitney U test. P values < 0·05 were considered significantly different.
Results
Iron status, faecal iron concentrations, weight gain and diet consumption
Fe status of rats fed different Fe diets was assessed by blood Hb concentration measurements at midpoint (week 15) and at endpoint (week 19), while ferritin concentration was measured at endpoint (Table 1). Although rats were fed a highly Fe-deficient diet (2·9 mg Fe/kg diet) for up to 16 weeks after an initial colonisation period during which they were fed a standard diet, Hb and ferritin concentrations at endpoint (week 19) were not significantly different between the groups of rats. However, lighter caecal content colour and liver colour were observed in the majority of rats in the Fe-deficient group (Fig. S1, available online). Weight gain was similar between all groups of rats during the first feeding period (weeks 3–15). During the second feeding period (weeks 15–19), rats in the control group exhibited the highest weight gain, which was significantly higher than that in rats in the 70 ppm Fe-supplemented group and Fe-excess group possibly because the control group had the lowest weight at week 15 and therefore more growth potential during the second feeding period. Interestingly, diet consumption was similar between the groups of rats during the first and second feeding periods.
Table 1 Hb and ferritin concentrations, weight gain and dietary intake in rats fed a diet differing only in iron concentration at midpoint (week 15) and endpoint (week 19) (n 7–8, each group) (Mean values with their standard errors)
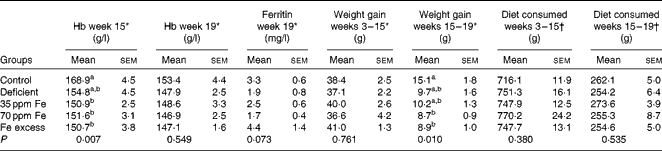
a,bMean values within a column with unlike superscript letters were significantly different (P< 0·05).
* Differences among groups were tested by ANOVA and post hoc Bonferroni test.
† Differences among groups were tested by non-parametric Kruskal–Wallis test.
At endpoint, Fe concentration in the faecal samples of rats in the Fe-deficient group (20 (sem 3) mg Fe/kg) was significantly lower than that in the faecal samples of rats in the 35 ppm Fe-supplemented group (185 (sem 14) mg Fe/kg, P< 0·001) and control group (184 (sem 7) mg Fe/kg, P< 0·001).
Effect of different iron concentrations in the diet on histological colitis scores
Ileum, caecum and colon of rats were collected after killing them, preserved in paraformaldehyde, and investigated by light microscopy for infiltration of immune cells, damage in crypt architecture, hyperaemia and mucosal erosion to evaluate histological colitis scores (Fig. 2(a)–(c)). In general, no severe colitis was detected in the ileum, caecum or colon of all rat groups, and the highest average score was 4·3 (sem 0·4) (ileum, 35 ppm Fe-supplemented group) of a maximum possible score of 11, indicating very light colitis. Rats fed an Fe-deficient diet had the lowest ileum histological colitis scores (score of 0·8 (sem 0·3)), which were significantly lower than those of the control rats (3·5 (sem 0·4), P= 0·001) and 35 ppm Fe-supplemented rats (4·3 (sem 0·4), P= 0·0009) (Fig. 2(a)). Caecum histological colitis scores were similar between all groups of rats and rats in the 35 ppm Fe-supplemented group had the highest scores (2·9 (sem 0·5)) (Fig. 2(b)). Supplementation with 35 ppm Fe after feeding an Fe-deficient diet (35 ppm Fe-supplemented group) significantly increased the caecum histological colitis scores compared with those of rats in the Fe-deficient group. Surprisingly, when a diet with 70 ppm Fe was fed after feeding an Fe-deficient diet (70 ppm Fe-supplemented group), no increase in histological colitis scores was observed. The opposite observation was made in the colon, wherein the 70 ppm Fe-supplemented group had higher histological colitis scores (4·0 (sem 0·4)) than the Fe-deficient group (Fig. 2(c)). Moreover, differences in histological colitis scores between the different parts of the intestine (ileum–caecum–colon) were observed with a lower histological colitis score being observed for the caecum in the control and 70 ppm Fe-supplemented groups and an increase in histological colitis scores from the ileum to the colon being observed in the Fe-deficient group. By contrast, a significant decrease in histological colitis scores from the ileum to the colon was observed in rats in the 35 ppm Fe-supplemented group, while no changes were observed in those in the Fe-excess group.

Fig. 2 Histological colitis scores of (a) ileum, (b) caecum and (c) colon of rats fed diets differing in iron concentration. Colitis scores were obtained by light microscopy after haematoxylin/eosin/safranin staining and investigation of infiltration of immune cells, damage in crypt architecture, hyperaemia and mucosal erosions. Each dot represents one rat. Horizontal bars are means, with their standard errors. * Mean value was significantly different from that of the control group (P< 0·05; non-parametric Mann–Whitney U test). † Mean value was significantly different from that of the iron-deficient group (P< 0·05; non-parametric Mann–Whitney U test).
Caecal and faecal microbiota composition in rats fed diets differing in iron concentration
Caecal contents of all rats were collected after killing them and the extracted DNA was investigated by qPCR to enumerate different bacterial groups present in the gut microbiota (Fig. 3) and by 454-pyrosequencing (Fig. 4(a)–(c)) to assess gut microbiota diversity and composition. qPCR did not reveal changes in the log number of total 16S rRNA gene copies or Firmicutes 16S rRNA gene copies/g caecum between the different groups of rats fed different Fe diets. The bacterial community in all rats was dominated by Firmicutes, including Clostridium cluster IV, followed by Bacteroides spp. (Fig. 3). This finding was confirmed by 454-pyrosequencing in rats in the control group, Fe-deficient group and 35 ppm Fe-supplemented group, where the phylum Firmicutes (54·5–82·6 %, data not shown) dominated, followed by Bacteroidetes (14·3–42·0 %, data not shown). On the taxonomic family level, these rats had the highest relative abundance (34·1–43·8 %) of Lachnospiraceae (Firmicutes) (Fig. 4(a)).

Fig. 3 Caecal microbiota composition after killing in rats fed diets differing only in iron concentration. Bacterial groups were enumerated with specific bacterial primers targeting the 16S rRNA gene or a specific functional gene by quantitative PCR. Values are means (n 8; iron excess, n 7), with their standard errors represented by vertical bars. a,b,cMean values within the same bacterial group with unlike letters were significantly different (P< 0·05; ANOVA followed by post hoc Bonferroni test). SRB, sulphate-reducing bacteria. ![]() , Control;
, Control; ![]() , Fe deficient;
, Fe deficient; ![]() , 35 ppm Fe;
, 35 ppm Fe; ![]() , 70 ppm Fe;
, 70 ppm Fe; ![]() , Fe excess.
, Fe excess.

Fig. 4 Caecal microbiota composition of rats fed the 35 ppm iron diet (n 3 samples), rats fed an iron-deficient diet (n 3 samples) and rats fed the control diet (n 3 samples). Relative abundances of (a) bacterial families and (b) genera identified by 454-pyrosequencing analysis. (c) Relative abundance of subdominant genera Bilophila, Coprococcus, Enterococcus and Turicibacter. Values are means, with their standard errors represented by vertical bars. * Mean values were significantly different from those of the iron-deficient group (P< 0·05; non-parametric Mann–Whitney U test). (a) ![]() , Verrucomicrobiaceae;
, Verrucomicrobiaceae; ![]() , Veillonellaceae;
, Veillonellaceae; ![]() , Alcaligenaceae;
, Alcaligenaceae; ![]() , Enterococcaceae;
, Enterococcaceae; ![]() , Desulfovibrionaceae;
, Desulfovibrionaceae; ![]() , Coriobacteriaceae;
, Coriobacteriaceae; ![]() , Erysipelotrichaceae;
, Erysipelotrichaceae; ![]() , Porphyromonadaceae;
, Porphyromonadaceae; ![]() , Rikenellaceae;
, Rikenellaceae; ![]() , Ruminococcaceae;
, Ruminococcaceae; ![]() , Clostridiaceae;
, Clostridiaceae; ![]() , Lachnospiraceae;
, Lachnospiraceae; ![]() , Bacteroidaceae;
, Bacteroidaceae; ![]() , smaller taxa;
, smaller taxa; ![]() , unclassified. (b)
, unclassified. (b) ![]() , Turicibacter;
, Turicibacter; ![]() , Desulfovibrio;
, Desulfovibrio; ![]() , Bilophila;
, Bilophila; ![]() , Sutterella;
, Sutterella; ![]() , Enterococcus;
, Enterococcus; ![]() , Alistipes;
, Alistipes; ![]() , Faecalibacterium;
, Faecalibacterium; ![]() , Clostridium;
, Clostridium; ![]() , Ruminococcus;
, Ruminococcus; ![]() , Coprococcus;
, Coprococcus; ![]() , Dorea;
, Dorea; ![]() , Blautia;
, Blautia; ![]() , Parabacteroides;
, Parabacteroides; ![]() , Bacteroides;
, Bacteroides; ![]() , smaller taxa;
, smaller taxa; ![]() , unclassified. (c)
, unclassified. (c) ![]() , Control;
, Control; ![]() , Fe deficient;
, Fe deficient; ![]() , 35 ppm Fe.
, 35 ppm Fe.
The levels of dominant bacterial groups (Firmicutes, Bacteroides spp. and Clostridium cluster IV) in caecum were similar in rats in the control and Fe-deficient groups. However, the levels of E. hallii and Enterobacteriaceae were significantly lower in rats fed an Fe-deficient diet (8·59 (sem 0·17) and 7·89 (sem 0·11), respectively; mean log 16S rRNA gene copies/g caecum) than in the control rats (9·22 (sem 0·08) and 8·61 (sem 0·16), respectively) (Fig. 3). Moreover, a significantly lower abundance of Bilophila spp. (0·36 (sem 0·11) %) and Coprococcus spp. (0·74 (sem 0·07) %) was observed in rats in the Fe-deficient group than in those in the control group (Bilophila spp., 0·87 (sem 0·19) %; Coprococcus spp. 2·11 (sem 0·58) %) by 454-pyrosequencing (Fig. 4(c)). Bacteroides spp., Clostridium cluster IV, F. prausnitzii, E. hallii and SRB were significantly increased in the caecum of rats in the 35 ppm Fe-supplemented group than in that of rats in the Fe-deficient group; however, bacterial concentrations did not increase to higher levels compared with those in the control group (Fig. 3). The 454-pyrosequencing analysis revealed a significantly higher relative abundance of Lachnospiraceae (43·81 (sem 2·79) %, Fe-deficient group: 34·14 (sem 1·97) %), Ruminococcaceae (12·15 (sem 1·30) %, Fe-deficient group: 8·25 (sem 0·38) %) and Veillonellaceae (0·46 (sem 0·05) %, Fe-deficient group: 0·29 (sem 0·02) %) in the 35 ppm Fe-supplemented group than in the Fe-deficient group (Fig. 4(a)). On the genus level, the abundance of Coprococcus spp. (2·21 (sem 0·43) %, Fe-deficient group: 0·74 (sem 0·07) %) and Bilophila spp. (0·77 (sem 0·05) %, Fe-deficient group: 0·36 (sem 0·11) %) was significantly increased, while that of Enterococcus spp. (0·05 (sem 0·03) %, Fe-deficient group: 1·24 (sem 1·03) %) and Turicibacter spp. (0·23 (sem 0·14) %, Fe-deficient group: 1·41 (sem 0·38) %) was significantly decreased in the 35 ppm Fe-supplemented group than in the Fe-deficient group (Fig. 4(b) and (c)). Bacterial populations in rats in the 70 ppm Fe-supplemented group and the Fe-excess group did not differ when compared with those in rats in the Fe-deficient group or control group, respectively (Fig. 3).
Faecal samples collected at baseline (week 3), midpoint (week 15) and endpoint (week 19) shortly before killing were analysed for microbial composition using qPCR to enumerate prevalent bacterial groups (Table S2, available online). During the entire trial period of 16 weeks, the faecal microbiota was very stable, with changes in the log numbers of 16S rRNA gene copies of dominant bacterial groups per g faeces being less than 1 log, and no direct correlations between dietary Fe concentrations and faecal microbiota composition could be extracted.
Effect of different iron diets on caecal microbiota metabolic activity
Fermentative metabolites and SCFA concentrations measured by HPLC (Fig. 5) were not different between rats in the Fe-deficient group and those in the control group. However, supplementation with Fe at the 35 and 70 ppm levels affected the caecal microbiota metabolic profile profoundly. Fermentative metabolites and acetate and propionate concentrations were all significantly increased in rats in the 35 ppm Fe-supplemented group (83·2 (sem 10·1), 33·7 (sem 2·3) and 15·3 (sem 0·9) mm, respectively) and the 70 ppm Fe-supplemented group (62·0 (sem 2·6), 33·1 (sem 1·6) and 14·5 (sem 0·9) mm, respectively) than in those in the Fe-deficient group (39·6 (sem 1·6), 23·7 (sem 1·2) and 9·5 (sem 0·5) mm, respectively). Moreover, butyrate concentration was significantly increased in the 35 ppm Fe-supplemented group (29·5 (sem 7·5) mm) compared with the Fe-deficient group (4·8 (sem 0·8) mm). Propionate and butyrate concentrations in the 35 ppm Fe-supplemented group were even higher than those in the control group. On the other hand, supplementation with 70 ppm Fe led to no significant increase in butyrate concentration compared with that in rats in the Fe-deficient group. Propionate (15·4 (sem 0·7) mm) and butyrate (16·7 (sem 3·9) mm) concentrations were significantly increased in the Fe-excess group compared with the control group (11·7 (sem 0·8) and 3·3 (sem 0·6) mm, respectively).

Fig. 5 Caecal fermentative metabolites and acetate, propionate and butyrate concentrations after killing in different groups of rats. Concentrations were measured by HPLC in caecum water samples in duplicate. Values are means (n 8; 35 ppm iron, n 7), with their standard errors represented by vertical bars. a,b,c,dMean values of the same metabolite with unlike letters were significantly different (P< 0·05; one-way ANOVA followed by post hoc Bonferroni test). ![]() , Fermentative metabolites;
, Fermentative metabolites; ![]() , acetate;
, acetate; ![]() , propionate;
, propionate; ![]() , butyrate.
, butyrate.
Discussion
The results of the present study carried out using human gut microbiota-associated rats provide a new insight into the highly complex interactions between the host, the gut microbiota and Fe. Colonisation of rats with human gut microbiota was successful, and dominant bacterial groups were present at similar levels in rats at baseline and in the donor microbiota as described previously( Reference Alpert, Sczesny and Gruhl 36 , Reference Crouzet, Gaultier and Del'homme 40 ). The use of one faecal donor also led to highly similar microbiotas in all rats, whereas in humans variability in microbiota composition between individuals is high and may potentially mask smaller effects( Reference Gootenberg and Turnbaugh 54 ). Feeding regimens were set up according to the classical Fe depletion–repletion study( Reference Forbes, Arnaud and Chichester 42 ) in which Fe depletion is started in very young rats (usually 3 weeks old), which have small body Fe stores and high Fe needs for growth, leading to severe Fe-deficiency anaemia( Reference Hilty, Arnold and Hilbe 28 , Reference Forbes, Arnaud and Chichester 42 ).
Surprisingly, in the present study, the 12- and 16-week depletion periods were insufficient to induce Fe-deficiency anaemia. This may be because the rats were 8 weeks of age when the intervention was started (instead of the usual 3 weeks) and had time to build up adequate Fe stores. Drawing on these stores allowed them to maintain Hb concentrations during the depletion period. However, rats in the Fe-deficient group had caecal contents of a lighter colour, suggesting a mild degree of Fe depletion (Fig. S1, available online). In addition, Fischer 344 rats are known to be more resistant to Fe deficiency than the Sprague–Dawley rats usually used in a Hb depletion–repletion study( Reference Kasaoka, Yamagishi and Kitano 55 ). At endpoint, faecal Fe concentrations in rats were higher than dietary Fe concentrations, which may be explained by a concentration effect of Fe in the gut lumen during water and nutrient absorption and a possible Fe loss in the host by sloughed enterocytes.
Dietary Fe supplementation has been shown to be associated with increased inflammatory reactions of the gut mucosa possibly generated by the production of reactive oxygen species in the presence of elevated luminal Fe concentration, which has been observed in inflammatory bowel disease patients and rat models( Reference Lih-Brody, Powell and Collier 56 – Reference Carrier, Aghdassi and Jeejeebhoy 58 ), while other studies suggest an involvement of gut bacterial composition changes due to Fe concentrations( Reference Werner, Wagner and Martinez 25 , Reference Perl, Fogarty and Harpaz 59 ). Werner et al. ( Reference Werner, Wagner and Martinez 25 ) found that a low-Fe diet completely inhibited gut inflammation in a mouse colitis model, while Fe supplementation in wild-type mice had no effect on colitis scores. In agreement, the present study showed lower histological colitis scores of ileum and colon in rats fed an Fe-deficient diet while Fe supplementation with 35 ppm Fe but not with 70 ppm Fe caused light colitis in ileum and caecum. Therefore, no clear inflammation pattern related to Fe concentrations could be detected in the investigated rats. Fe supplementation in school children in Côte d'Ivoire has been found to clearly increase calprotectin concentrations, but in an environment with a high risk of pathogen exposure( Reference Zimmermann, Chassard and Rohner 30 ). We speculate that Fe supplementation in the range of physiological levels may not lead to mucosal inflammation unless other influencing factors are present such as a high contamination with pathogens or an inflammatory pre-set of the host.
Faecal microbiota composition in all rats was very stable over time. Because rats had normal Fe status, we speculated that especially under an Fe-deficient diet regimen, bacteria in the colon could sequester Fe from sloughed enterocytes. Therefore, caecal contents were analysed, in which Fe might have a more direct impact on the microbiota. There was no difference in gut microbiota metabolic activity in caecum between rats in the Fe-deficient group and those in the control group. However, we found that low luminal Fe concentrations slightly affected the gut microbiota composition and decreased the relative abundances of Bilophila spp., E. hallii and Coprococcus spp. most probably due to Fe-dependent enzymes such as hydrogenases in their metabolic pathways( Reference Pryde, Duncan and Hold 9 , Reference da Silva, Venceslau and Fernandes 60 , Reference Calusinska, Happe and Joris 61 ).
The findings of the present study are in contrast to previous findings in highly Fe-deficient rats and also to the results of an in vitro colonic fermentation experiment( Reference Dostal, Chassard and Hilty 24 , Reference Dostal, Fehlbaum and Chassard 27 ). In both experiments, very-low-Fe conditions had caused major dysbiosis of the gut microbiota and especially decreased butyrate and propionate production. Moreover, it has been shown that changes in host Fe metabolism in mice and humans have effects on the gut microbiota without changing dietary Fe concentrations( Reference Buhnik-Rosenblau, Moshe-Belizowski and Danin-Poleg 32 , Reference Balamurugan, Mary and Chittaranjan 62 ). However, in the present study, hosts were not Fe deficient and it is possible that host Fe plays an important role in the maintenance of the composition and metabolic function of the gut microbiota as part of the symbiosis between the microbiota and the host. Indeed, during low-Fe diet feeding, faecal Fe concentrations were 20 mg Fe/kg faeces, which could be provided by unabsorbed dietary Fe and by sloughed enterocytes from the host. These faecal Fe concentrations are probably adequate to maintain the gut microbiota as has been shown in our previous in vitro studies( Reference Dostal, Fehlbaum and Chassard 27 ).
Fe supplementation with 35 mg Fe/kg diet from FeSO4 (35 ppm Fe-supplemented group) had a strong effect on both the caecal microbiota composition and metabolic activity, especially increasing the abundance of Bacteroides spp. and butyrate producers such as Clostridium cluster IV members, e.g. F. prausnitzii, and Coprococcus spp. Moreover, a decrease in the relative abundances of Turicibacter spp. and Enterococcus spp., both of which are opportunistic pathogens( Reference Cuiv, Klaassens and Durkin 63 , Reference Arias and Murray 64 ), was observed during feeding of the 35 ppm Fe diet than during feeding of a Fe-deficient diet. Enterococci most probably had a growth advantage under low-Fe conditions due to their restricted need for Fe and lost this advantage during Fe supplementation. Similar observations were made in mice in which feeding a diet containing Fe led to an increase in the abundance of Bacteroides spp. and to a decrease in that of Turicibacter spp. compared with feeding a Fe-free diet( Reference Werner, Wagner and Martinez 25 ).
The concentrations of acetate, propionate and especially butyrate were significantly increased due to Fe supplementation compared with those in rats in the Fe-deficient group. In a previous study, Fe supplementation has been found to lead to a significant increase in butyrate and propionate concentrations in highly Fe-deficient rats( Reference Dostal, Chassard and Hilty 24 ). Moreover, Fe-deficient conditions in the same rat study and also in a colonic in vitro fermentation study led to a decrease in butyrate and propionate production( Reference Dostal, Fehlbaum and Chassard 27 ). These observations suggest that Fe is a crucial element for butyrate and propionate production in strict anaerobic gut bacteria. Indeed, in the butyrate production pathway, oxidoreductases and hydrogenases are involved( Reference Louis and Flint 8 , Reference De Vuyst and Leroy 14 , Reference Louis, Young and Holtrop 65 ), which are often Fe dependent, and under conditions of optimal bioavailable Fe concentrations, the conversion of dietary or host carbons into end metabolites such as butyrate by gut bacteria may be enhanced. Studies in bioreactors with mixed strict-anaerobe cultures have shown that increasing Fe concentrations lead to a higher H2 and butyrate yield per carbon source( Reference Lee, Shinde and Choi 29 , Reference Karadag and Puhakka 66 , Reference Wei, Dong and Wang 67 ). Indeed, when calculating the total carbon output from bacterial fermentation in the form of SCFA, the diet with 35 mg Fe/kg diet from FeSO4 (35 ppm Fe-supplemented group) led to more than double the carbon output than the control diet or a Fe-deficient diet, mainly due to the promotion of butyrate production, although diet consumption was similar in these groups of rats. SCFA can provide up to an additional 10 % of daily dietary energy to the host from indigestible compounds, such as fibres( Reference Conterno, Fava and Viola 68 ). This promotion of carbon output due to Fe supplementation could increase the energy source of plant-based diets, which are mainly consumed in developing countries, and therefore might contribute to weight gain in malnourished individuals.
A significant increase in caecal butyrate and propionate concentrations along with a decrease in the abundance of possible opportunistic pathogens due to 35 ppm Fe supplementation could lead to beneficial effects on host gut health. Propionate produced by several Bacteroides spp. has been shown to be involved in the regulation of satiety( Reference Hosseini, Grootaert and Verstraete 69 ). Butyrate is the main energy source for colonocytes, and it can inhibit NF-κB activation and therefore decrease inflammatory responses( Reference Luhrs, Kudlich and Neumann 70 , Reference Luhrs, Gerke and Muller 71 ). Moreover, butyrate has anti-carcinogenic effects through the promotion of apoptosis and inhibition of proliferation( Reference Roy, Dionne and Marx 72 ). Interestingly, no correlation between increased butyrate concentrations and decreased colitis scores was observed in the present study, but the anti-inflammatory effects of butyrate might be much more visible in mucosa with an inflammatory pre-set. Moreover, the abundance of H2-utilising and potentially toxic H2S-producing SRB was increased with Fe supplementation in the present study possibly due to the promotion of the production of H2, a by-product of the butyrate production pathway( Reference Louis, Young and Holtrop 65 ). SRB have been identified as possible contributors of different digestive pathologies such as inflammatory bowel syndrome and inflammatory bowel disease( Reference Crouzet, Gaultier and Del'homme 40 , Reference Chassard, Dapoigny and Scott 73 – Reference Attene-Ramos, Nava and Muellner 76 ), and H2S has been shown to affect metabolic functions in colonocytes and to cause DNA damage( Reference Carbonero, Benefiel and Alizadeh-Ghamsari 74 , Reference Attene-Ramos, Nava and Muellner 76 ). However, other studies have suggested a protective role of H2S in the epithelial layer during inflammation and identified H2S as an important mediator for intracellular processes( Reference Wallace, Vong and McKnight 77 , Reference Hirata, Naito and Takagi 78 ). Therefore, it remains difficult to directly associate an increase in SRB counts and H2S production with negative effects on the gut mucosa, and no negative modulation of the gut mucosa could be detected in the present study.
In conclusion, the present study carried out using human gut microbiota-associated rats investigated the effects of dietary Fe concentrations on the gut microbiota. Our data suggest that an Fe-deficient diet alone may have no major effects on dominant bacterial populations or gut microbiota metabolic activity in a host, which is not Fe deficient based on blood parameters as observed in the study. By contrast, Fe supplementation with 35 mg Fe/kg diet from FeSO4 promoted dominant bacterial groups and slightly increased SRB, while the abundance of potential opportunistic pathogens was decreased. Moreover, Fe supplementation strongly increased the metabolic activity of the gut microbiota. Histological colitis scores remained very low despite Fe supplementation, indicating that Fe alone does not lead to gut inflammation. Thus, we suggest that Fe supplementation might confer additional health benefits on the host by stimulating the gut microbiota without an inflammatory gut mucosa and in a relatively pathogen-free environment.
Supplementary material
To view supplementary material for this article, please visit http://dx.doi.org/10.1017/S000711451400021X
Acknowledgements
The authors thank Christophe De Martrin and Gérard Vert for their assistance in animal care and maintenance as well as Rainer Follador (Microsynth AG) for bioinformatics analysis of the pyrosequencing data.
The present study was funded by grants from the Swiss National Science Foundation (project number: 310030_127272, Bern, Switzerland) and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (award number: U01HD0 64921). The Swiss National Science Foundation and the Eunice Kennedy Shriver National Institute of Child Health and Human Development had no role in the design and analysis of the study or in the writing of this article.
The authors' contributions are as follows: A. D., C. C., A. B.-D., C. D. and C. L. designed the study; A. D., C. C., C. D. and V. T. P. carried out the study; A. D. and V. T. P. analysed the data; A. D. wrote the manuscript; C. C., A. B.-D., C. D., M. B. Z. and C. L. edited the manuscript.
None of the authors has any conflicts of interest to declare.








