Published online by Cambridge University Press: 02 April 2024
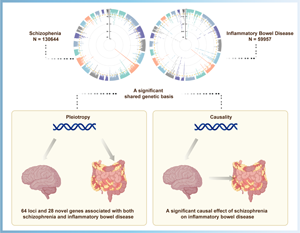
The comorbidity between schizophrenia (SCZ) and inflammatory bowel disease (IBD) observed in epidemiological studies is partially attributed to genetic overlap, but the magnitude of shared genetic components and the causality relationship between them remains unclear.
By leveraging large-scale genome-wide association study (GWAS) summary statistics for SCZ, IBD, ulcerative colitis (UC), and Crohn's disease (CD), we conducted a comprehensive genetic pleiotropic analysis to uncover shared loci, genes, or biological processes between SCZ and each of IBD, UC, and CD, independently. Univariable and multivariable Mendelian randomization (MR) analyses were applied to assess the causality across these two disorders.
SCZ genetically correlated with IBD (rg = 0.14, p = 3.65 × 10−9), UC (rg = 0.15, p = 4.88 × 10−8), and CD (rg = 0.12, p = 2.27 × 10−6), all surpassed the Bonferroni correction. Cross-trait meta-analysis identified 64, 52, and 66 significantly independent loci associated with SCZ and IBD, UC, and CD, respectively. Follow-up gene-based analysis found 11 novel pleiotropic genes (KAT5, RABEP1, ELP5, CSNK1G1, etc) in all joint phenotypes. Co-expression and pathway enrichment analysis illustrated those novel genes were mainly involved in core immune-related signal transduction and cerebral disorder-related pathways. In univariable MR, genetic predisposition to SCZ was associated with an increased risk of IBD (OR 1.11, 95% CI 1.07–1.15, p = 1.85 × 10−6). Multivariable MR indicated a causal effect of genetic liability to SCZ on IBD risk independent of Actinobacteria (OR 1.11, 95% CI 1.06–1.16, p = 1.34 × 10−6) or BMI (OR 1.11, 95% CI 1.04–1.18, p = 1.84 × 10−3).
We confirmed a shared genetic basis, pleiotropic loci/genes, and causal relationship between SCZ and IBD, providing novel insights into the biological mechanism and therapeutic targets underlying these two disorders.
Jing Wang, Guang-Yu Luo, and Tian Tian contributed equally to this paper.