Ornithine aminotransferase (OAT; l-ornithine 2-oxo acid aminotransferase; EC 2.6.1.13) is a pyridoxal-5′-phosphate-dependent mitochondrial matrix aminotransferase that catalyses the interconversion of ornithine into glutamate semi-aldehyde, which is in spontaneous equilibrium with its cyclic tautomer, pyrroline-5-carboxylate (P5C). In contrast to other aminotransferases, this reaction enables the reversible transfer of the ornithine ω-amino group. It depends on two tyrosine residues leading to the interaction with ornithine and the protection of the ω-amino group against hydrolysis(Reference Markova, Peneff, Hewlins, Schirmer and John1).
OAT is localised at a crossing between two important metabolisms, with arginine and polyamine metabolism on one side and glutamate and proline metabolism on the other (Fig. 1).

Fig. 1 Overview of the arginine (Arg)–glutamine (Gln) pathway (adapted from Dekaney et al. (Reference Dekaney, Wu and Jaeger50)). OAT, ornithine aminotransferase; Orn, ornithine; Glu, glutamic acid; Cit, citrulline; AS, argininosuccinate. Note that no tissue expressed all of the enzymes.
Although OAT is expressed constitutively in many tissues, it is mainly found in the liver, kidney (at higher levels in females) and the small intestine(Reference Herzfeld and Knox2, Reference Wu, Davis, Flynn, Knabe and Davidson3) where its response to hormones and variations in dietary protein intake is subject to complex regulation mechanisms(Reference Herzfeld and Knox2, Reference Boon, Geerts, Jonker, Lamers and Van Noorden4, Reference O'Sullivan, Brosnan and Brosnan5). The reaction is directed towards glutamate semi-aldehyde synthesis in the liver and kidneys and towards ornithine synthesis in the small intestine. Hepatic OAT expression is restricted to perivenous hepatocytes(Reference Boon, Geerts, Jonker, Lamers and Van Noorden4) and co-localised with glutamine synthetase expression(Reference O'Sullivan, Brosnan and Brosnan6), which suggests that OAT plays a major role in N homeostasis. In the kidney, Levillain et al. (Reference Levillain, Diaz, Reymond and Soulet7) showed that OAT is distributed along the whole nephron, but its activity is higher in proximal tubules than in distal tubules. In the intestine, OAT is expressed mainly in the villous epithelium of the jejunum and more weakly in the crypt epithelium(Reference Ozaki, Gotoh, Nagasaki, Miyanaka, Takeya, Fujiyama, Tomita and Mori8).
OAT does not appear to exist in different isoenzymic forms: Sanada et al. (Reference Sanada, Suemori and Katunuma9) showed that hepatic, renal and intestinal OAT share the same physico-chemical and immunochemical properties. In addition, intestinal and hepatic OAT display similar kinetic properties(Reference Herzfeld and Raper10).
OAT activity has been shown to be tightly regulated, with important tissue-specific differences. The murine, rat and human OAT gene presents a high degree of homology with a cAMP response element binding protein responsive element in the − 134 to − 122 region and several AGGTCA-like motifs in the promoter ( − 222 to − 205 and − 366 to − 396) which correspond to the consensus binding site for various members of the nuclear receptor family(Reference Shull, Esumi, Colwell, Pennington and Jendoubi11). As suggested by the cAMP response element binding protein site, there is cAMP-dependent regulation of OAT expression, but it is restricted to the liver where OAT is induced by glucagon, and this effect is inhibited by glucose(Reference Merrill, Mueckler and Pitot12–Reference Fagan, Lazaris-Karatzas, Sonenberg and Rozen14). In the kidney, OAT expression is decreased by testosterone(Reference Manteuffel-Cymborowska, Peska, Chmurzynska and Grzelakowska-Sztabert15, Reference Levillain, Diaz, Blanchard and Dechaud16) and induced by oestradiol(Reference Herzfeld and Knox2, Reference Sanada, Suemori and Katunuma9, Reference Fagan, Lazaris-Karatzas, Sonenberg and Rozen14, Reference Patnaik, Sarangi and Patnaik17) and thyroid hormones(Reference Fagan, Lazaris-Karatzas, Sonenberg and Rozen14, Reference Mueckler and Pitot18, Reference Mueckler, Moran and Pitot19).
Besides age-related variations(Reference Patnaik, Sarangi and Patnaik17, Reference Patnaik and Patnaik20, Reference Volpe, Sawamura and Strecker21), protein supply is also a key regulator of hepatic and intestinal (but not renal) OAT activity(Reference Bourdel, Hitier, Lardeux and Girard-Globa22, Reference Matsuzawa, Kobayashi, Tashiro and Kasahara23). A protein load is followed by a rapid and marked induction of hepatic OAT, with normalisation in nearly 12 h(Reference Matsuzawa, Kobayashi, Tashiro and Kasahara23). This effect occurs at transcriptional(Reference Boon, Geerts, Jonker, Lamers and Van Noorden4, Reference Mueckler and Pitot18), translational(Reference Mueckler and Pitot18, Reference Burcham, Giometti, Tollaksen and Peraino24) and post-translational levels(Reference Chee and Swick25). Conversely, a protein-restricted diet doubles OAT activity in the small intestine(Reference Matsuzawa, Kobayashi, Tashiro and Kasahara23). This could be either a non-specific N-related effect or may be related to specific amino acids(Reference Volpe, Sawamura and Strecker21, Reference Civen, Brown and Trimmer26, Reference Peraino, Blake and Pitot27).
Given that OAT is the unique metabolic pathway linking arginine and glutamine (which are two major players involved in N homeostasis), these data collectively suggest that OAT could play an important role in the regulation of glutamate and ornithine availability, and thus glutamine and arginine metabolism. The importance of OAT at the whole-body level has mostly been demonstrated for the neonatal period, since OAT-null mice generated to study gyrate atrophy die 24–48 h after birth if they do not receive arginine supplementation(Reference Wang, Lawler, Steel, Sipila, Milam and Valle28). Interestingly, these mice presented increased ornithine concentration at the whole-body level, likewise in OAT inhibition experiments, suggesting the reaction favours ornithine consumption at the whole-body level. However, it is still unknown to what extent OAT influences amino acid metabolism and N homeostasis in adults.
Our working hypothesis is that OAT contributes both to the adaptation to the level of protein supply and to the regulation of the availability of arginine and glutamine, two amino acids that play a key role in N homeostasis(Reference Cynober, Lennarz and Lane29).
To answer this question, we generated a model of transgenic mice overexpressing OAT in the three main organs expressing this enzyme, i.e. the liver, the intestine and the kidney. We compared the kinetic properties of endogenous and overexpressed enzymes and evaluated the effect of OAT overexpression on N metabolism.
The present paper reports that OAT overexpression does not induce any phenotypic alterations in adult mice, but only a decrease in hepatic ornithine concentrations, meaning that overexpression of OAT does not seem to influence amino acid homeostasis.
Materials and methods
Chemicals
Chemicals were purchased from Sigma Aldrich (Saint Quentin Fallavier, France), Virbac (Carros, France), Pfizer (Paris, France), Invitrogen (Cergy-Pontoise, France) and Bio-Rad (Marne-la-Coquette, France), except where otherwise indicated.
Generation of the transgenic mice
The EAB-9K-OAT transgene was produced by inserting, at the EcoRV site, the blunted NotI-SalI fragment of the human OAT (hOAT) cDNA into the plasmid construct containing the chimeric EAB-9K promoter construct which is known to allow expression of the transgene in liver, intestine and kidney(Reference Cadoret, Ovejero, Saadi-Kheddouci, Souil, Fabre, Romagnolo, Kahn and Perret30). The chimeric EAB-9K promoter contains the 4·9 kbp regulatory sequences (from − 4580 to +365 of the rat calbindin-D9K gene)(Reference Colnot, Romagnolo, Lambert, Cluzeaud, Porteu, Vandewalle, Thomasset, Kahn and Perret31) linked to the enhancer EAB of the rat aldolase B gene(Reference Gregori, Porteu, Mitchell, Kahn and Pichard32). This promoter was chosen as the best compromise between tissue specificity, as it targets the three main organs of arginine metabolism (i.e. liver, kidney and intestine), and strength. The hOAT cDNA containing the 1320 bp open reading frame of the hOAT gene was obtained from a clone IMAGE kindly provided by the RZPD (German Resource Centre for Genome Research). Transgenic mice (EAB-9K-OAT) were obtained by microinjecting fertilised (C57/BPXDBA) F1 mouse eggs with the EAB-9K-OAT hybrid DNA construct. Transgenic mice were identified by Southern blotting (data not shown). We identified one founder that produced one transgenic line that was then genotyped by PCR using the following primers: forward 5′-GAGGGTAGGGGGATCGG and reverse 5′-CAATGCCCTTGGTTGACAGC. The study was conducted on heterozygous mice and wild-type (WT) mice of the same litter were used as controls.
Animals and feeding conditions
Animal care followed French and European Community guidelines for the protection of animals used for experimental and other scientific purposes. Mice were kept in standard cages and given ad libitum access to a standard regimen (M20; Dietex, Saint-Gratien, France).
Experimental design
Fifteen WT (seven males and eight females; aged 3 months) C57/BL6 mice and fifteen C57/BL6 transgenic mice (OAT; seven males and eight females; aged 3 months) were used. The animals were anaesthetised by intraperitoneal injection of 10 ml/kg of a solution containing ketamine (100 mg/ml; Virbac) and medetomodin (1 mg/ml; Domitor®; Pfizer) and killed by cervical dislocation. Blood was removed by intracardiac puncture by a first operator. Blood samples were collected in heparin tubes (Starstedt, Orsay, France) for amino acid quantification. Blood samples were centrifuged immediately for 10 min at 4°C (5000 rpm) and the plasma was deproteinised with sulfosalicylic acid (30 mg/ml). After a 10 min period of incubation at 4°C, the samples were centrifuged for 10 min at 4°C (5000 rpm), and the supernatant fractions were collected, frozen in liquid N2 and stored at − 80°C until amino acid determination.
Quickly after blood removal, a second operator made a laparotomy, and promptly removed 10 cm of proximal jejunum from each animal. Jejunal section samples were washed with ice-cold saline solution flushed through the lumen and then inverted to collect the mucosa using glass scrapers. Three sections of proximal jejunum mucosa were collected. Simultaneously, a third operator collected three samples of liver that were weighed and stored at − 80°C for quantification of tissue amino acids and RNA and assay of OAT activity. The same operator also removed both kidneys that were weighed and stored at − 80°C for mRNA expression analysis and assay of OAT activity.
Amino acid concentrations
The frozen tissues were homogenised in ice-cold 10 % TCA containing 0·5 mm-EDTA and 200 μm-norvaline (internal standard). The acid-soluble fraction containing free amino acids was separated from precipitated proteins by centrifugation (10 min at 4°C and 2500 g). The supernatant fractions were stored at − 80°C until amino acid analysis. Amino acids in tissues and deproteinised plasma were separated and quantified by ion-exchange chromatography with spectrophotometric detection after ninhydrin derivatisation using an amino acid analyser (AminoTac JLC-500/V; Jeol, Tokyo, Japan)(Reference Neveux, David, Cynober and Cynober33). Results are expressed as nmol/g tissue (wet weight) or μmol/l plasma.
Ornithine aminotransferase activity
OAT activity was measured in liver, jejunum and kidney homogenates as described by Herzfeld & Knox(Reference Herzfeld and Knox2). Tissues were homogenised (100 mg/ml) at 4°C in a homogenisation buffer (0·33 m-sucrose, 5 mm-HEPES, 1 mm-ethylene glycol tetra-acetic acid, 1 mm-dithiothreitol, 0·5 % Triton X-100, pH 7·4) using an Ultra-Turrax (Ika-Labortechnik, Staufen, Germany) homogeniser. In order to disrupt mitochondria, the homogenates were subjected to three cycles of freezing in liquid N2 and thawing at 37°C, and then centrifuged at 600 g for 10 min. The assays were performed with different quantities of supernatant fractions depending on the organ (25 μl for liver, 50 μl for kidney and 100 μl for jejunum) that were added to 200 μl of an assay mixture consisting of 75 mm-potassium phosphate buffer (pH 7·5), 20 mm-ornithine, 0·45 mm-pyridoxal phosphate, 5 mm-o-aminobenzaldehyde, and 0 or 3·75 mm-α-ketoglutarate, and incubated for 15 min at 37°C. OAT activity was quantified by measuring the condensation product of P5C with o-aminobenzaldehyde by spectrophotometry at 440 nm. The amount of P5C produced was calculated based on its molar extinction coefficient of 2·71 × 103/m per cm(Reference Strecker34). Enzyme activity is presented as mmol P5C formed per min at 37°C and expressed as a ratio to the amount of protein.
The kinetic properties of OAT were determined on liver samples from both WT and transgenic mice at six different concentrations of ornithine (from 5 to 30 mm) and of α-ketoglutarate (from 0·615 to 2 mm), using an incubation time of 45 min. Double reciprocal plots of the rates measured and secondary graphs plotted for each substrate allowed us to obtain maximum rate of reaction (V max) and K m data.
Tissue proteins were quantified using the Biuret method (BC kit; Uptima-Interchim, Montluçon, France). After homogenisation, 25 μl of samples (diluted at 1:50 in sterile water) were placed in duplicate into microplates. Then 200 μl of reagent mix were added to each well, and the plates were incubated for 30 min at 37°C. Protein concentration was determined spectrophotometrically at 550 nm. Bovine albumin was used as standard.
Immunohistochemistry
One mouse from each group was killed by cervical dislocation. Liver samples were embedded in paraffin for histological and immunocytochemical analysis, as previously described(Reference Colnot, Niwa-Kawakita, Hamard, Godard, Le Plenier, Houbron, Romagnolo, Berrebi, Giovannini and Perret35). Sections (5 μm) were cut off the paraffin-embedded liver and treated with 3 % H2O2 and an avidin–biotin blocking kit (Vector Laboratories, Burlingame, CA, USA).
Non-specific sites were coated with PBS containing 100 mm-phosphate buffer, 150 mm-NaCl, supplemented with 0·3 % Triton X-100 and 1 % bovine serum albumin (PBST-BSA) for 120 min and incubated overnight at 4°C with purified primary rabbit anti-glutamine synthetase antibody (dilution 1:200) in PBST. The slides were rinsed three times in PBST for 5 min and incubated for 120 min with secondary Alexa Fluor 546-conjugated goat anti-rabbit IgG (dilution 1:1000; Interchim, Montluçon, France). Slides were washed twice with 4′,6-diamidine-2-phenylindole dihydrochloride (0·1 μg/ml) in PBS for 30 min. Slides were washed three times in PBS and mounted with Fluoprep (Biomérieux, Marcy l'Etoile, France). Tissue sections were examined with a fluorescent microscope.
In situ hybridisation
Immediately after killing the mouse, the liver and the entire gastrointestinal tract were removed. The liver was fixed in 10 % formol. The intestine was splayed open along its length, and then rolled up from the proximal to distal end to form a Swiss roll. In situ hybridisation was carried out on 7 μm slices of the paraffin-embedded Swiss rolls. The 201 bp digoxigenin-labelled RNA probe recognising murine OAT (mOAT) (fragment from 1333 to 1466 nucleotides) and hOAT (fragment from 1247 to 2009 nucleotides) was prepared by in vitro transcription with the digoxigenin RNA labelling kit (Roche, Meylan, France) using T7 RNA polymerases. Sections were incubated overnight at 68°C in the prehybridisation buffer containing 200 ng/ml of digoxigenin-labelled RNA probe. Immunodetection of the hybridised probe was carried out using an anti-digoxigenin antibody (1:4000; Roche).
mRNA quantification
Total RNA was extracted from mouse tissues using Trizol reagent (Invitrogen, Cergy-Pontoise, France) according to the manufacturer's protocol. Messenger RNA were analysed by real-time PCR. Reverse transcription was performed with 2 μg of total RNA using the Superscript™ First-Strand Synthesis System for RT-PCR (Invitrogen). mRNA quantification was performed on a SmartCycler System (Cepheid, Sunnyvale, CA, USA) with a 1:100 dilution of the cDNA using the SYBRGreen Kit for SmartCycler (Eurogentec, Angers, France). One of the experimental samples was selected as the reference, the basis sample (i.e. as the 100 % value). For all experimental samples, target quantity was divided by the reference quantity to generate the relative levels of expression (as described in Applied Biosystems: Guide to Performing Relative Quantification of Gene expression Using Real-Time Quantitative PCR, 2004). Thus, the amount of target is a unitless number and all quantities are expressed as an n-fold difference relative to the reference sample. Serial dilutions of the reference of liver, kidney or jejunum were made from WT mice for mOAT and from the kidney from a transgenic mouse for hOAT expression. Quantitative values were obtained from the threshold cycle number (Ct) at which the increase in growth of PCR product can be detected. Standard curves were generated by linear regression using Ct v. log dilution. The equivalent amounts of mOAT and hOAT cDNA were calculated from Ct values using the standard curves. Ct values varied between twenty-five and twenty-seven cycles for mOAT and twenty-three and twenty-eight cycles for hOAT in transgenic mice, and exceeded thirty-two cycles for WT mice. Data were expressed as the ratio between the target gene and the internal control (18S), thus giving relative expression. Specific primer sequences were as follow: mOAT, forward primer 5′-GGGCTCTTGTGAAACTCTGC and reverse primer 5′-AGATGGGTCCGTTTCTCCTT; hOAT, forward primer 5′-AGACTGCCTGTAAACTAGCTCGTAAG and reverse primer 5′-ACTGGAGATAGCAGACAACGTCCT; 18S, forward primer 5′-GTAACCCGTTGAACCCCATT and reverse primer 5′-CCATCCAATCGGTAG(Reference Ovejero, Cavard and Périanin36).
Statistics
Results are presented as mean values with their standard errors. Statistical analysis was performed on Statview 5.0 software (StataCorp LP, College Station, TX, USA) using two-way ANOVA followed by a Newman–Keuls test, and the sort criteria were sex and genotype. A probability < 0·05 was considered as significant.
Results and discussion
We used the chimeric EAB-9K promoter construct to target expression of the hOAT gene in the liver, kidney and intestine (Fig. 2 (A)). The efficiency of these regulatory sequences has already been demonstrated for the expression of an oncogenic mutant β-catenin cDNA(Reference Cadoret, Ovejero, Saadi-Kheddouci, Souil, Fabre, Romagnolo, Kahn and Perret30). We first analysed expression of the transgene by in situ hybridisation. The probe used allowed us to analyse both the expression of the hOAT transgene and the mouse endogenous OAT, due to similarity between the two species. As expected, we showed in the WT animals an expression of the endogenous OAT restricted to the perivenous area of the liver, in hepatocytes that co-expressed the glutamine synthetase gene (Fig. 2 (B i and ii)). Regarding transgenic mice, we observed an extension of the OAT expression towards the periportal area, in hepatocytes that do not express the glutamine synthetase gene (Fig. 2 (B iii and iv)). In the intestine, we observed a very weak expression of the endogenous gene in the epithelial cells of the small intestine villi in WT animals (Fig. 2 (B v)). Expression of OAT was strongly increased in transgenic animals and was also restricted to the epithelial cells of the villi (Fig. 2 (B vi)).

Fig. 2 Expression of ornithine aminotransferase (OAT) in wild-type (WT) and transgenic mice. (A) Diagram of the EAB/9K–hOAT construct. (B) In situ hybridisation (i, iii, v and vi) of OAT in liver and intestine, and immunohistocheminal detection (ii and iv) of OAT in the liver of WT and transgenic mice. Expression of OAT is confined to perivenous hepatocytes that expressed the glutamine synthetase gene in WT mice while the area of expression was extended towards the periportal area in hepatocytes that do not express the glutamine synthetase gene. In the intestine, overexpressed and endogenous OAT is localised in epithelial cells along the villi. (i) to (iv), Liver samples; (i) and (ii), WT mice; (iii) and (iv), transgenic mice; (v) and (vi), intestinal samples; (v), WT mice; (vi), transgenic mice.
Expression of the hOAT transgene was then analysed by real-time RT-PCR to study the difference in expression levels among the three organs targeted by the tissue-specific promoter. Transgene expression was found stronger in the liver and the kidney than in the jejunum of transgenic mice (Table 1). This may be explained by the presence in the 5′-flanking sequence of an oestrogen-responsive element in the rat CaBP9K promoter used in the construction of the tissue-specific promoter(Reference L'Horset, Blin, Colnot, Lambert, Thomasset and Perret37, Reference Romagnolo, Cluzeaud, Lambert, Colnot, Porteu, Molina, Tomasset, Vandewalle, Kahn and Perret38). However, transgene expression in the liver was greater in males than in females. Yet, this cannot fully explain the difference between males and females and may point to other regulatory processes.
Table 1 Amounts of human ornithine aminotransferase (hOAT) in the liver, kidney and jejunum of transgenic mice*
(Mean values with their standard errors)
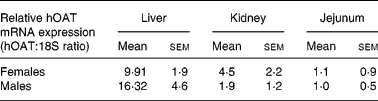
* hOAT mRNA was measured by RT-PCR in liver, kidney and jejunum samples taken from transgenic mice.
Glutamine has a number of important regulatory roles, increasing protein synthesis and decreasing protein degradation, for example(Reference Smith39), and arginine functions as a secretagogue stimulating the release of growth hormone, insulin-like growth factor 1, insulin and prolactin(Reference Palmer, Walter and Ensinck40, Reference Rakoff, Siler, Sinha and Yen41). Therefore, it can be hypothesised that a modification in glutamine or arginine availability could lead to changes in protein metabolism and thus in body composition. However, the mice overexpressing OAT did not show phenotypic modifications in body weight, organ weight, protein content (Table 2), growth rate, food consumption or life span compared with WT female and male mice of the same litter. This is at variance with another enzyme of arginine metabolism, arginase I, whose overexpression in murine enterocytes induces marked alterations (hair and muscle growth retardation and lymphoid tissue development) and significant metabolic changes, including arginine deficiency(Reference de Jonge, Hallemeesch and Kwikkers42).
Table 2 Organ and body weights, protein content, and number of wild-type (WT) and transgenic mice
(Mean values with their standard errors)

OAT, ornithine aminotransferase.
Moreover, our data indicate that the expression of endogenous mOAT is not modified by the overexpression of hOAT (Fig. 3).

Fig. 3 Basal expression of murine ornithine aminotransferase (mOAT) in the liver, kidney and jejunum of wild-type (□) and transgenic (■) female (A) and male (B) mice. Expressions were determined by RT-PCR. Results are relative to 18S as an internal control. Data are means, with standard errors represented by vertical bars.
As a consequence of its overexpression, OAT activity was significantly higher in the liver and the female kidney but not in the intestine (Fig. 4). It should be underlined that in transgenic mice, OAT activity corresponds to the total activity of both mOAT and hOAT. However, given the similar kinetic parameters (K m and V max) between WT and transgenic mice, it can reasonably be assumed that the enzymes from the two species share similar kinetic properties. hOAT and endogenous mOAT displayed similar kinetic profiles, with no differences between the apparent OAT K m and V max values for the two genotypes for both substrates (Fig. 5). It should be highlighted that the apparent K m for α-ketoglutarate was more than fivefold lower than that for ornithine, showing that overexpressed hOAT, like mOAT(Reference Burcham, Giometti, Tollaksen and Peraino24), has a higher affinity for α-ketoglutarate. Thus, the highly conserved OAT DNA sequence(Reference Dzikowska, Swianiewicz, Talarczyk, Wisniewska, Goras, Scazzocchio and Weglenski43) would lead to highly similar enzymic properties, in agreement with previous literature data(Reference Ohura, Kominami, Tada and Katunuma44, Reference Lim, Rho, Park, Jhee, Kim and Kim45).

Fig. 4 Effect of ornithine aminotransferase (OAT) overexpression on OAT enzymic activity in the liver, kidney and jejunum (nmol/μg protein) of wild-type (□) and transgenic (■) female (A) and male (B) mice. Data are means, with standard errors represented by vertical bars. * Mean value was significantly different from that of the female mice (P < 0·05). † Mean value was significantly different from that of the WT mice (P < 0·05).

Fig. 5 Secondary graphs of intercepts from Lineweaver–Burk plots for the substrates α-ketoglutarate (A) and ornithine (B) (mmol) and ornithine aminotransferase (OAT) kinetic values in wild-type (WT; ■) and transgenic (OA; ●) mice. Values were estimated from Lineweaver–Burk plots of rates and secondary graphs of intercepts. Reaction rates were measured over a concentration range of 0·615 to 2 mm for α-ketoglutarate and 5 to 30 mm for ornithine in liver samples. Velocity is expressed as μmol product formed/min per mg tissue. The experiment was repeated three times. A typical result is presented here. V max, maximum rate of reaction.
In addition, OAT activity in the kidney (Fig. 4) was greater in females than in males. This finding is in agreement with a previous study(Reference Levillain, Diaz, Reymond and Soulet7) showing that OAT expression is induced by oestrogens in rats.
The increase in OAT activity is associated with only limited modifications in plasma and tissue amino acids (Table 3). The most prominent modification is a decrease in ornithine levels in the liver and the intestine, without any significant difference for other amino acids and total amino acid content in the tissues under study.
Table 3 Effect of ornithine aminotransferase (OAT) overexpression on plasma, liver and jejunum concentrations of selected amino acids†
(Mean values with their standard errors)

WT, wild type; ND, non-detectable.
* Mean value was significantly different from that for the WT mice (P < 0·05).
† Results are expressed for n 6–8 per group.
The decrease in hepatic ornithine is in agreement with a previous report(Reference O'Sullivan, Brosnan and Brosnan6) indicating that hepatic metabolism is oriented towards ornithine utilisation for glutamate synthesis. The OAT expression in periportal hepatocytes would be consistent with an enhanced consumption of ornithine in the liver, which would then be converted into glutamate. This could also deplete ornithine available for the urea cycle. Although urinary urea determination would have been a better indicator for demonstrating variations in urea cycle, it is not easily achievable in mice in individual metabolism cages. However, first there was no difference in plasma phenylalanine (data not shown), which is considered a suitable marker of protein turnover and has been shown to correlate negatively with N excretion(Reference Coudray-Lucas, Cynober, Lioret, Saizy, Baux and Giboudeau46). Moreover, glutamate and urea plasma levels (data not shown) were similar between WT and transgenic mice, even though plasma urea is more a reflection of renal function than liver production.
In the absence of modifications in plasma arginine levels in OAT-overexpressing mice, the low ornithine levels in the liver of these mice is unlikely to be due to alterations in arginase activity. Indeed, previous work on genetically modified arginase-knock-out or arginase-overexpressing mice highlighted marked alterations in plasma and tissue arginine concentrations. Experiments with [13C]ornithine and isolated perfused mouse liver are required to evaluate more precisely the impact of OAT overexpression in periportal hepatocytes. The stability of the glutamate pool may result from various different mechanisms: (i) the difference between the pool sizes of ornithine and glutamate, since glutamate content is roughly four-fold larger than that of ornithine, which means that the mean 80 μm decrease in ornithine would lead to only a limited increase in the glutamate pool; (ii) ornithine is not the sole contributor to the glutamate pool; (iii) glutamate produced by OAT may be readily used for glutamine synthesis. Indeed, the co-localisation of OAT with glutamine synthetase in perivenous hepatocytes(Reference Kuo, Hwu, Valle and Darnell47) is further evidence for a role of these cells in scavenging ammonia which could have escaped elimination by ureagenesis in periportal cells(Reference Haussinger, Graf and Weiergraber48). Finally, it is difficult to see any OAT overexpression-induced changes in liver and plasma glutamine levels, as glutamine is the major interorgan N transporter in mice and has a very large plasma pool.
\Interestingly, ornithine concentrations in the jejunum were also lower in transgenic mice (Table 3), despite no significant modification in OAT activity, although the transgene was effectively expressed. However, ornithine accumulation in situations of OAT inhibition implies that, at the whole-body level, the reaction equilibrium favours ornithine utilisation, to form proline or glutamine, and thus does not only depend on OAT activity in the gut. Since this result cannot be due to OAT overexpression in the jejunum, it must be an indirect consequence of the OAT overexpression in the liver or kidney depleting ornithine pools and therefore decreasing ornithine availability for the intestine in the post-absorptive state. However, this observation suggests that a modification in amino acid pools in physiological conditions may lead to significant modifications in amino acid homeostasis in specific tissues, even if plasma levels of ornithine and related metabolites are not modified. Future studies to determine the precise contribution of OAT overexpression to amino acid metabolism will require interorgan flux measurement(Reference Hallemeesch, Soeters and Deutz49) using stable isotopes and arteriovenous difference measurements.
Thus, the results obtained in the present study suggest that as far as tissue and plasma amino acid concentrations are concerned, OAT overexpression in the liver and the kidney seems to induce only slight changes in N metabolism in healthy mice.
Since the most significant property of OAT is its place at the crossroads of various amino acid metabolisms, other enzymes involved in these same metabolic pathways (for example, arginase, glutamine synthetase) may have to compensate for modifications in amino acid concentrations in order to restore N homeostasis. It would be of interest to cross our OAT-overexpressing mice with mice carrying genetically modified expressions of one or several other enzymes in this crossroad, such as arginase, in order to ascertain whether the whole metabolic pathway would be affected in the absence of compensation. Moreover, the influence of OAT overexpression on the response to variations in dietary protein content, and/or to a challenge to N homeostasis (for example, sepsis) could be studied to give vital clues as to the real contribution of OAT to N homeostasis.
In conclusion, to the best of our knowledge, this is the first study to have created and used a viable model of mice overexpressing OAT mainly in the liver and the kidney, and to a lesser extent in the small intestine, in order to better define the role of the OAT enzyme in N homeostasis. The model highlighted an inverse relationship between OAT activity and hepatic ornithine concentration.
Acknowledgements
G. V. collected the data, analysed the data and wrote the manuscript. All the other authors read the manuscript and contributed to the discussion. J.-P. D. B., C. M. and L. C. designed the study and supervised the project. F. S. helped design the experiment, collect the data and analyse the data. C. P. generated the transgenic mice model and provided significant advice. D. R. designed the OAT kinetics study, helped collecting the data and analysed the data. S. L. P. and C. G. contributed to the design of the experiments and provided excellent technical support.
Our work was supported by funds from the Ministry of Research and Technology (EA2498). No other financial or contractual agreements might cause conflicts of interest or be perceived as causing conflicts of interest. There are no financial arrangements between an author and a company whose product figures prominently in the paper.










