Cholesterol is an essential component of cell membranes and also serves as a precursor for life-sustaining steroid hormones and bile acids( Reference Grundy 1 ). It is well known that deregulation of cholesterol metabolism contributes to obesity, diabetes and CVD( Reference Grundy 2 , Reference Grundy, Cleeman and Daniels 3 ). Moreover, cholesterol is particularly essential for embryogenesis( Reference Woollett 4 , Reference Roux, Wolf and Mulliez 5 ), and low plasma cholesterol level is usually correlated with low body weight at birth( Reference Edison, Berg and Remaley 6 – Reference Lu, Pond and Mersmann 8 ).
Hepatic cholesterol homeostasis is maintained through the coordinated regulation of three relevant processes: biosynthesis; transportation; transformation( Reference Woollett 4 , Reference Khovidhunkit, Kim and Memon 9 ). In particular, sterol regulatory element-binding protein-2 (SREBP2)( Reference Horton, Shimomura and Brown 10 ) and 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR)( Reference Brown and Goldstein 11 ) are key factors/enzymes for cholesterol biosynthesis, LDL receptor (LDLR) and HDL receptor (scavenger receptor class B type I (SR-BI)) are responsible for transportation( Reference Wyne and Woollett 12 ), while cholesterol-7α-hydroxylase (CYP7α1) and cholesterol-27α-hydroxylase (CYP27α1)( Reference Dias and Ribeiro 13 ) are main enzymes catalysing transformation to bile acids. It has been shown that hepatic cholesterol homeostasis in offspring is highly vulnerable to maternal nutrition status( Reference Sohi, Marchand and Revesz 14 , Reference Goharkhay, Tamayo and Yin 15 ), predominantly through epigenetic mechanisms such as DNA methylation, histone modification and microRNA-mediated post-transcriptional regulation( Reference Cong, Jia and Li 16 ). Methyl donors, such as methionine or folic acid, are able to reverse the epigenetic modifications and thereby restore the behavioural or metabolic disorders in offspring caused by prenatal or neonatal insults( Reference Weaver, Champagne and Brown 17 , Reference Lillycrop, Rodford and Garratt 18 ).
Betaine, also known as betaine anhydrous or trimethylglycine, functions as a substrate for the formation of methionine, which can then be further converted to S-adenosylmethionine (SAM)( Reference Lever and Slow 19 ). In the methionine metabolic cycle, SAM is further converted to S-adenosylhomocysteine by glycine N-methyltransferase (GNMT), thereby donating the methyl group for DNA and protein methylation catalysed by DNMT (DNA methyltransferases) and histone methyltransferases, respectively. Consequently, betaine supplementation may modulate gene expression through modifying epigenetic marks such as DNA and histone methylation( Reference Ji, Nordgren and Chai 20 , Reference Bestor 21 ).
Betaine deficiency results in a series of metabolic abnormalities( Reference Lever and Slow 19 ), while betaine supplementation in the diet can prevent obesogenic diet-induced hepatic steatosis in rodents( Reference Cordero, Gomez-Uriz and Campion 22 ) and improve growth performance and carcass characteristics in domestic animals( Reference Eklund, Bauer and Wamatu 23 , Reference Ratriyanto, Mosenthin and Bauer 24 ). It has been observed that betaine supplementation could change cholesterol production in human subjects and animals( Reference McGregor, Dellow and Robson 25 – Reference Martins, Neves and Freitas 28 ), yet the mechanisms remain unclear. Furthermore, although betaine is known to be critical for embryonic and fetal development( Reference Lever and Slow 19 ), the effects of betaine supplementation in maternal diet during gestation on hepatic cholesterol metabolism in neonatal offspring and the underlying mechanisms have not been investigated.
In the present study, we aimed to investigate the effects of feeding gestational sows with a betaine-supplemented diet on hepatic cholesterol metabolism in newborn piglets. To explore the potential epigenetic mechanisms underlying such effects, we studied hepatic expression of genes involved in cholesterol and methionine metabolism, and determined the status of DNA and histone methylation on the promoter of cholesterol metabolic genes, as well as the expression of microRNA targeting these genes, in the liver of neonatal piglets.
Materials and methods
Animals and sampling
A total of sixteen second-parity cross-bred sows (Landrace × Yorkshire) at the age of approximately 8 months were artificially inseminated, at the observation of oestrus, with a mixture of Duroc semen samples obtained from two littermate boars. The sows were randomly divided into control and betaine groups (eight per group) after 1 week of the artificial insemination. The sows in the control group were fed a basal diet not supplemented with betaine, while those in the betaine group received a betaine-supplemented (3 g/kg) diet throughout pregnancy. The composition of the two experimental diets is shown in Table 1. Betaine (Skystone Feed Company Limited) was added in the form of betaine hydrochloride with 98 % purity. All the sows were fed three times per d at 05.00, 10.00 and 17.00 hours, and had free access to water. Immediately after parturition, newborn piglets were individually weighed and the piglets from the same litter were kept together in a warm creep area. Male piglets (one per litter) of the mean body weight of 1·59 kg were selected and killed before suckling. Serum samples were prepared immediately, and the liver (without the gall bladder) was taken within 20 min postmortem, snap-frozen in liquid N2 and stored at − 80°C until further analysis.
Table 1 Composition and nutrient content of the experimental diets

* The premix contained the following vitamins and minerals (per kg of diet): vitamin A, 72 mg; vitamin D3, 1·5 mg; vitamin E, 648 mg; vitamin K3, 30 mg; vitamin B1, 30 mg; vitamin B2, 120 mg; vitamin B6, 60 mg; vitamin B12, 360 mg; niacin, 600 mg; pantothenic acid, 300 mg; folic acid, 6 mg; manganese sulphate, 1·0 g; zinc oxide, 2·5 g; ferrous sulphate, 4·0 g; copper sulphate, 4·0 g; sodium selenite, 6 mg; Ca, 150 g; P, 15 g; NaCl, 40 g.
The experimental protocol was approved by the Animal Ethics Committee of Nanjing Agricultural University, with the project number 2012CB124703. The slaughter and sampling procedures complied with the ‘Guidelines on Ethical Treatment of Experimental Animals’ (2006) No. 398 set by the Ministry of Science and Technology, China.
Determination of hepatic betaine and S-adenosylmethionine content and serum methionine concentration
For the determination of betaine concentrations, 1 g of frozen liver samples was prepared as described previously( Reference Kirsch, Herrmann and Rabagny 29 ). Betaine concentrations in the liver samples were measured with a liquid chromatography (Agilent 1200; Agilent Technologies)–MS (API 5000TM; AB Sciex) system.
Hepatic SAM content was measured using the HCB Quantitative Porcine Competitive ELISA kit (S200FC; Hermes Criterion Biotechnology) following the manufacturer's instructions. Serum concentration of methionine was determined in duplicate with an automatic amino acid analyser (L-8900; Hitachi).
Measurement of cholesterol and bile acid concentrations
Serum concentration of total cholesterol was measured using a commercial cholesterol assay kit (E1005; Applygen Technologies, Inc.). Serum concentrations of LDL-cholesterol (LDL-C) and HDL-cholesterol were measured with assay kits (006340 and 006328, respectively; Beijing BHKT Clinical Reagent Company Limited). Hepatic total cholesterol concentration was measured using a tissue total cholesterol assay kit (E1015; Applygen Technologies, Inc.) following the manufacturer's instructions. Hepatic total bile acid concentrations were determined by enzymatic colorimetric methods using a commercial kit (E003; Nanjing Jiancheng Bioengineering Institute).
Real-time PCR for mRNA quantification
For RNA extraction, approximately 200 mg of liver samples were homogenised in 1 ml of TRIzol reagent (Invitrogen), according to the manufacturer's protocol. RNA samples (2 μg) were digested with DNase and reverse transcribed to complementary DNA using random hexamer primers (Promega). Diluted complementary DNA (2 μl, 1:25) was used in each real-time PCR assay with Mx3000P (Stratagene). Peptidylprolyl isomerase A (PPIA) was chosen as a reference gene to normalise the mRNA abundance of target genes, because it is expressed in comparable abundance to target genes and was not affected by the experimental factor. Real-time PCR data were analysed by the 2− ΔΔCt method. Primers for real-time PCR were synthesised by Generay Biotech and are listed in online supplementary Table S2.
Protein extraction and Western blot analysis
Total cellular protein and nuclear protein were extracted from 200 mg of frozen liver samples as described previously( Reference Liu, Wang and Li 30 ). Protein concentrations were measured with a Pierce BCA Protein Assay kit (no. 23 225; Thermo Scientific). Western blot analysis of target proteins was carried out according to the protocols provided by the manufacturer. The sources of primary antibodies used in Western blot are listed in online supplementary Table S3. GAPDH or β-actin was selected as the loading controls for total cellular protein, while histone H1 (H1) was selected as the loading control for nuclear proteins.
DNA methylation analysis – methylated DNA immunoprecipitation
Genomic DNA extracted from the liver samples was sonicated to produce small fragments ranging from 300 to 1000 bp. The fragmented DNA (2 μg) was heat-denatured to produce single-stranded DNA. A mouse monoclonal antibody against 5-methyl cytidine (ab10805; Abcam) was used to immunoprecipitate the methylated DNA fragments. Pretreated protein G agarose beads (40 μl, 50 % slurry, sc-2003; Santa Cruz Biotechnology) were used to capture the immune complexes. The beads bound to immune complexes were washed to eliminate non-specific binding and resuspended in 250 μl digestion buffer containing proteinase K to purify the immunoprecipitated methylated DNA. A small aliquot of immunoprecipitated methylated DNA was used to amplify the proximal promoter sequences of target genes by real-time PCR. A pair of negative control primers was used to amplify a promoter region absent of CpG sites as the internal control. The results of methylated DNA immunoprecipitation were calculated relative to the internal control, and expressed as the fold change relative to the mean value of the control group. The specific and negative control primers are listed in online supplementary Table S2.
Chromatin immunoprecipitation assay
Approximately 200 mg of frozen liver samples were ground in liquid N2 and resuspended with PBS containing protease inhibitor cocktail (no. 11697498001; Roche). Cross-linking of protein and DNA was performed by adding formaldehyde to a final concentration of 1 %, and then the reaction was terminated with glycine (2·5 mol/l) at room temperature. The reaction mix was centrifuged and the pellets were rinsed with PBS and homogenised in a SDS lysis buffer containing protease inhibitors. Crude chromatin preparations were sonicated to an average length ranging from 200 to 500 bp and precleared with salmon sperm DNA-treated protein G agarose beads (40 μl, 50 % slurry, sc-2003; Santa Cruz Biotechnology). The mixture of precleared chromatin preparations and 2 μg of specific primary antibody were incubated overnight at 4°C (for details of antibodies, see online supplementary Table S2). A negative control was included with normal rat IgG. Protein G agarose beads (40 μl, 50 % slurry, sc-2003; Santa Cruz Biotechnology) were added to capture the immunoprecipitated chromatin complexes. Finally, DNA fragments were released from the immunoprecipitated complexes by reverse cross-linking at 65°C for 1 h, and quantitative real-time PCR was used to quantify the fragments of target gene promoters with specific primers (see online supplementary Table S2) using purified immunoprecipitated DNA as the template.
Quantification of microRNA
Total RNA (2 μg) treated with RNase-free DNase I (Promega) were polyadenylated by poly(A) polymerase using a poly(A) tailing kit (AM1350; Applied Biosystems), according to the manufacturer's instructions. Polyadenylated RNA was then dissolved and reverse transcribed using a poly(T) adapter. Real-time PCR was performed with SYBR Green qPCR master mix reagent (Takara) in triplicate using a microRNA (miRNA)-specific forward primer and a universal reverse primer complementary to part of the poly(T) adapter sequence. U6 small-nuclear RNA (U6 snRNA) was used as a reference gene to normalise the expression of miRNA.
The sequences of all porcine miRNA were acquired from miRBase (http://www.mirbase.org/). miRNA targeting HMGCR, LDLR, CYP7α1 and CYP27α1 were predicted with an online miRNA prediction tool( Reference Kertesz, Iovino and Unnerstall 31 ). Among all the predicted miRNA, four miRNA targeting HMGCR, ten targeting LDLR, five targeting CYP7α1 and twelve targeting CYP27α1 were quantified by real-time PCR. The primer sequences used for miRNA analysis are listed in online supplementary Table S4.
Functional validation of microRNA
Genomic sequences of porcine miR-497 and miR-181 precursors (see online supplementary Table S5) were synthesised and inserted into the pSilencer 3.0-H1 small-interfering RNA expression vector by Invitrogen. The 3′-UTR (untranslated regions) of the porcine HMGCR gene containing a conserved motif for miR-497 targeting was amplified by PCR using the primers 5′-GGGCGGGGGGACCAGCCTGCTCCCTCA-3′ and 5′-GGAGAGAAGCACGCTCCGCTGGAGG-3′. The 3′-UTR of the CYP7α1 gene containing a conserved motif for miR-181 targeting was amplified using the primers 5′- TCTGACATTACTAAACATGCCTCTA-3′ and 5′- TAACATTTAAAACATAATAAATTTA-3′. The PCR products were then cloned downstream to the pGL3-Control luciferase reporter vector (Promega) and formed the plasmids pGL3-Control/HMGCR and pGL3-Control/CYP7α1, respectively.
HeLa cells were cultured in Dulbecco's modified Eagle's medium containing 10 % fetal bovine serum at 37°C in a 5 % CO2 incubator, and co-transfected with 100 ng pGL3-Control/HMGCR, 10 ng Herpes simplex virus-thymidine kinase promoter (pRL-TK) plasmid and 100 ng pSilence 3·1 H1-neo miR-497, or with 100 ng pGL3-Control/CYP7α1, 10 ng pRL-TK and 100 ng pSilencer 3·1 H1-neo miR-181, for 24 and 48 h, respectively. Luciferase activity of the cells was measured on a luminometer according to the manufacturer's instruction using the luciferase reporter assay system (GloMax® 96 Microplate Luminometer; Promega).
Statistical analysis
All data are presented as means with their standard errors. Comparisons were made using independent two-tailed Student's t test with SPSS 17.0 for Windows (SPSS, Inc.). Results from the relative quantification of mRNA, protein, CpG methylation and miRNA are expressed as the fold change relative to the mean value of the control group. Differences were considered statistically significant at P< 0·05.
Results
Reproductive performance of sows and body weight and liver weight of piglets
Maternal betaine supplementation did not affect the litter size or litter weight (see online supplementary Table S1). Moreover, newborn piglets born to betaine-supplemented sows exhibited comparable body weight and liver weight to their control counterparts (Table 2).
Table 2 Body and liver weights and hepatic total cholesterol and bile acid content, and serum cholesterol concentration in newborn piglets (Mean values with their standard errors, n 8)
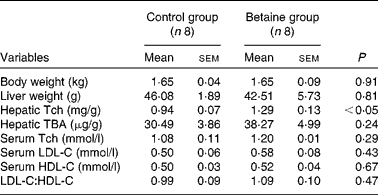
Tch, total cholesterol; TBA, total bile acids; LDL-C, LDL-cholesterol; HDL-C, HDL-cholesterol.
Hepatic betaine and methionine metabolism
Piglets born to betaine-supplemented sows had significantly higher hepatic betaine content (P< 0·05; Fig. 1(a)) and serum methionine concentration (P< 0·05; Fig. 1(b)). Moreover, hepatic SAM content tended to be higher (P= 0·09) in piglets born to betaine-supplemented sows (Fig. 1(c)). These alterations were associated with significantly up-regulated mRNA (P< 0·05; Fig. 1(d)) and protein (P< 0·05; Fig. 1(e) and (f)) expression of GNMT. However, no significant alteration was observed for the mRNA or protein expression of DNMT1, DNMT3a (P= 0·06) or DNMT3b in the liver of newborn piglets born to betaine-supplemented mothers (Fig. 1(d)–(f)).

Fig. 1 Hepatic betaine (a), serum methionine concentration (b) and hepatic S-adenosylmethionine (SAM) (c) content, gene (d) and protein (e, f) expression of glycine N-methyltransferase (GNMT) and DNA methyltransferases (DNMT) in the liver of newborn piglets. Values are means (n 8), with their standard errors represented by vertical bars. * Mean value was significantly different from that of the control group (P< 0·05). □, Control group; ■, betaine group.
Serum cholesterol concentrations and hepatic total cholesterol content
No significant alterations were observed in the serum concentrations of LDL-C- and HDL-cholesterol or in the ratio of LDL-C:HDL-cholesterol. However, hepatic content of total cholesterol was significantly increased (P< 0·05) in piglets born to betaine-supplemented sows (Table 2).
Hepatic expression of cholesterol metabolic genes
Among the genes involved in cholesterol de novo synthesis, HMGCR was significantly reduced (P< 0·05) by 45 % at the mRNA level in piglets born to betaine-supplemented sows (Fig. 2(a)), which was accompanied by a significant down-regulation (P< 0·05) of SREBP2 at both mRNA and nuclear protein content levels (Fig. 2(a)–(c)). However, the hepatic protein content of HMGCR was significantly higher (P< 0·05) in piglets born to betaine-supplemented sows (Fig. 2(b) and (c)).

Fig. 2 Hepatic mRNA abundance, Western blot analysis and summary of the protein expression of sterol regulatory element-binding protein-2 (SREBP2) and 3-hydroxy-3-methylglutaryl CoA reductase (HMGCR) in cholesterol biosynthesis (a–c), those of LDL receptor (LDLR) and scavenger receptor class B type I (SR-BI) in cholesterol transport and uptake (d–f), and those of cholesterol-7α-hydroxylase (CYP7α1) and cholesterol-27α-hydroxylase (CYP27α1) (g–i) in cholesterol transformation in the liver of newborn piglets. Values are means (n 8), with their standard errors represented by vertical bars. * Mean value was significantly different from that of the control group (P< 0·05). H1, Histone H1; GAPDH, glyceraldehyde 3-phosphate dehydrogenase. □, Control group; ■, betaine group.
Among the genes involved in cholesterol transport and uptake, although maternal betaine supplementation did not alter the serum concentrations of LDL-C or HDL-cholesterol in neonatal offspring (Table 2), hepatic LDLR was significantly down-regulated (P< 0·05) at both mRNA (Fig. 2(d)) and protein (Fig. 2(e) and (f)) levels. In contrast, neither the mRNA nor the protein expression of SR-BI was altered in betaine-exposed piglets (Fig. 2(d)–(f)).
Of the genes involved in cholesterol transformation, hepatic CYP7α1 mRNA was significantly up-regulated (P< 0·05) by 87 % in newborn piglets born to betaine-supplemented sows (Fig. 2(g)), but no alteration was observed at the protein level (Fig. 2(h) and (i)). In contrast, the expression of CYP27α1 was up-regulated (P< 0·05), at both mRNA and protein levels, in the liver of piglets born to betaine-supplemented sows (Fig. 2(g)–(i)).
Epigenetic modifications of altered cholesterol metabolic genes
The level of CpG methylation on the HMGCR promoter was 2·34-fold higher (P< 0·05) in the liver of piglets born to betaine-supplemented sows (Fig. 3(a)), which was in parallel with a 40 % increment (P< 0·05) of the repressive histone mark H3K27me3 (histone H3 lysine 27 trimethylation; Fig. 3(b)). Significant DNA hypermethylation (P< 0·05; Fig. 3(c)) and a trend of more enriched H3K27me3 mark (P= 0·06; Fig. 3(d)) was also detected in the promoter of the LDLR gene in the liver of piglets born to betaine-supplemented sows. Because no CpG islands were found within the 5′ flanking sequence of the porcine CYP7α1, CYP27α1 or SREBP2 genes, and the SR-BI mRNA level was not changed in betaine-exposed piglets, we excluded CYP7α1, CYP27α1, SREBP2 and SR-BI genes from methylated DNA immunoprecipitation and chromatin immunoprecipitation analyses in the present study.

Fig. 3 Methylated DNA immunoprecipitation (MeDIP) and chromatin immunoprecipitation (ChIP) analyses of DNA methylation and histone modifications on the gene promoter of 3-hydroxy-3-methylglutaryl CoA reductase (HMGCR) (a, b) and LDL receptor (LDLR) (c, d) in the liver of newborn piglets. Values are means (n 8), with their standard errors represented by vertical bars. * Mean value was significantly different from that of the control group (P< 0·05). H3, Histone H3; H3K4me3, histone H3 lysine 4 trimethylation; H3K27me3, histone H3 lysine 27 trimethylation. □, Control group; ■, betaine group.
Hepatic expression of microRNA targeting altered cholesterol metabolic genes
Piglets born to betaine-supplemented sows demonstrated a significant down-regulation (P< 0·05) in the hepatic expression of miR-497 (Fig. 4(a)), which is predicted to target HMGCR. Moreover, LDLR-targeting miRNA, including miR-1285, miR-138, miR-4334-5p and miR-7144-5p, were all up-regulated (P< 0·05) in the liver of betaine-exposed piglets (Fig. 4(b)). Furthermore, miR-181 predicted to target CYP7α1 was remarkably up-regulated (P< 0·05; Fig. 4(c)), while miR-17-3p, miR-202-3p, miR-30-3p, miR-361-3p and miR-7143-3p predicted to target CYP27α1 were significantly down-regulated (P< 0·05, Fig. 4(d)) in the liver of piglets born to betaine-supplemented sows.

Fig. 4 MicroRNA (miRNA) predicted to target 3′-UTR of 3-hydroxy-3-methylglutaryl CoA reductase (HMGCR) (a), LDL receptor (LDLR) (b), cholesterol-7α-hydroxylase (CYP7α1) (c) and cholesterol-27α-hydroxylase (CYP27α1) (d) in the liver of newborn piglets. Values are means (n 8), with their standard errors represented by vertical bars. * Mean value was significantly different from that of the control group (P< 0·05).
Functional validation of miR-497 and miR-181
Ectopic expression of miR-497 significantly suppressed (P< 0·05) the luciferase activity of HeLa cells co-transfected with the pGL3-Control/HMGCR luciferase reporter plasmid at both 24 and 48 h (Fig. 5(a)). Similarly, the luciferase activity of HeLa cells transfected with the pGL3-Control/CYP7α1 reporter plasmid was significantly decreased by co-transfection of pSilencer 3·1 H1-neo miR-181 (P< 0·05) at both 24 and 48 h (Fig. 5(b)).

Fig. 5 Validation of ssc-miR-497 (Sus scrofa miR-497) targeting 3-hydroxy-3-methylglutaryl CoA reductase (HMGCR) 3′-UTR (a) and ssc-miR-181 (Sus scrofa miR-181) targeting cholesterol-7α-hydroxylase (CYP7α1) 3’-UTR (b). Values are means (n 6), with their standard errors represented by vertical bars. * Mean value was significantly different from that of the control group (P< 0·05). (a) □, miRNA-sc (scrambled control); ■, miR-497. (b) □, miRNA-sc (scrambled control); ■, miR-181.
Discussion
In the present study, maternal betaine supplementation during gestation caused a significant increase in hepatic betaine content in neonatal offspring. In mammals, betaine is used as a substrate for methionine synthesis( Reference Dominguez-Salas, Moore and Cole 32 ). Dietary betaine supplementation has been shown to increase serum methionine concentrations in human subjects( Reference Storch, Wagner and Young 33 ). In agreement with these previous findings, we found elevated serum methionine concentrations associated with higher hepatic betaine content in betaine-exposed piglets. Previous observation indicates that excessive methionine leads to an accumulation of SAM and the activation of GNMT( Reference Varela-Rey, Martinez-Lopez and Fernandez-Ramos 34 ). Indeed, betaine-exposed piglets demonstrated a higher hepatic SAM content and activated GNMT expression. GNMT is known to provide methyl groups for DNA and protein methylation through the conversion of SAM to S-adenosylhomocysteine( Reference Cook and Wagner 35 ). Therefore, maternal betaine supplementation during gestation may modify the methylation of DNA and proteins, especially histones, in the liver of betaine-exposed piglets.
Dietary methionine supplementation has been shown to elevate cholesterol concentrations in the liver and plasma of rats( Reference Hirche, Schroder and Knoth 36 ). In the present study, neonates born to betaine-supplemented dams had 37·1 % higher hepatic cholesterol content, associated with altered expression of cholesterol metabolic genes involved in cholesterol biosynthesis, transformation and transportation. To our surprise, we found that mRNA abundances of SREBP2, LDLR and CYP27α1 were in accordance with their respective protein content, while HMGCR and CYP7α1 demonstrated dissociated mRNA and protein levels. The regulation of gene expression is known to occur at different levels, namely transcription, post-transcription and translation. For some genes, the regulation occurs predominantly at the level of transcription. In this case, mRNA abundance usually agrees with the protein content. However, many other genes are also subjected to post-transcriptional and/or translational regulations, and in which case, mRNA abundance and protein content can be uncoupled. mRNA abundance reported herein represent the steady-state mRNA level resulting from a balance between transcription and degradation. Proteins are considered to be the executors of biological functions. However, many proteins, especially enzymes, have to be modified post-translationally to gain biological activities. In the present study, neither mRNA abundance nor protein content is directly relevant to biological functions. The results presented herein only demonstrate that cholesterol metabolic genes are differently regulated at both transcription and/or post-transcription levels in the liver of betaine-exposed piglets.
DNA methylation and histone modifications are recognised as crucial mechanisms involved in the transcriptional regulation of cholesterol metabolic genes. For instance, maternal dietary protein restriction increased hepatic cholesterol content in rat offspring through modulating histone modifications on the CYP7α1 promoter and thus altering the mRNA expression of CYP7α1 ( Reference Sohi, Marchand and Revesz 14 ). Moreover, dietary protein restriction in sows increased hepatic HMGCR expression, which was associated with DNA hypomethylation and lower H3K27me3 on the HMGCR gene promoter( Reference Cong, Jia and Li 16 ). In the present study, the promoters of HMGCR and LDLR genes were found to be hypermethylated in the liver of piglets prenatally exposed to betaine, which coincided with the down-regulation of these two genes at the mRNA level. Furthermore, H3K27me3 is a repressive histone mark that negatively regulates transcription by promoting a compact chromatin structure( Reference Ringrose, Ehret and Paro 37 ). The diminished HMGCR and LDLR gene expression was associated with more enriched H3K27me3 on the respective gene promoters. Therefore, it appears that increased DNA methylation and repressive histone mark play a role in the transcriptional regulation of HMGCR and LDLR genes in the liver of piglets in response to prenatal betaine exposure.
It is well known that SREBP-2 is a major transcriptional factor that activates the expression of HMGCR and LDLR genes( Reference Horton, Shimomura and Brown 10 , Reference Goldstein and Brown 38 ). We found a significant down-regulation of SREBP-2 at both mRNA and protein levels in the liver of the betaine-exposed group, which appears to be the direct cause for repressed transcription of HMGCR and LDLR genes. It should be mentioned that the CpG islands on HMGCR and LDLR gene promoters detected in the present study are predicted to contain binding sites for a number of transcriptional factors including SREBP-2 (see online supplementary Fig. S1). This implicates that chromatin remodelling caused by altered DNA and histone methylation status on HMGCR and LDLR promoters may affect the binding of specific transcriptional factors, thereby regulating the transcriptional level of these genes.
Another interesting finding of the present study is the incongruity between the mRNA and protein levels for HMGCR and CYP7α1 genes. HMGCR was decreased at the mRNA level but increased at the protein level, while CYP7α1 was up-regulated at the mRNA level but remained unchanged at the protein level. The dissociation of mRNA abundance and protein content implies the possible involvement of post-transcriptional regulation. MicroRNA are known to participate predominantly in the post-transcriptional regulation through target mRNA degradation and/or translational repression. Previous studies have indicated that miRNA predominantly promotes the cleavage of mRNA in plants( Reference Kawasaki and Taira 39 , Reference Moxon, Jing and Szittya 40 ), while miRNA acts mainly through translational repression in animals( Reference Williams 41 ). Moreover, it has been reported that in animal models and cell lines, miRNA targets, in principal, protein translation rather than mRNA degradation( Reference Pan, Zheng and Zhao 42 , Reference Sangokoya, Doss and Chi 43 ). Therefore, when transcriptional regulation and miRNA-mediated translational repression are not synchronised, mRNA and protein levels can be uncoupled. In the present study, the down-regulated expression of miR-497 was associated with increased protein content of HMGCR, and the up-regulated expression of miR-181 was associated with the unchanged protein level of CYP7α1, in the liver of betaine-exposed piglets. To verify the targeting sites of miR-497 and miR-181 on 3′-UTR of HMGCR and CYP7α1 transcripts, respectively, we conducted luciferase reporter gene assay and confirmed that miR-497 and miR-181 inhibited luciferase activity through targeting 3′-UTR of HMGCR and CYP7α1, respectively.
Betaine insufficiency is associated with the metabolic syndrome including lipid disorder and diabetes, while betaine supplementation increases serum total cholesterol and LDL-C concentrations( Reference McGregor, Dellow and Robson 25 , Reference Schwab, Torronen and Toppinen 26 , Reference Rabinowitch 44 ). Betaine, as a methyl donor, is known to be critical for human embryonic and fetal development( Reference Lever and Slow 19 ), and plasma betaine concentration has been reported to be correlated with total cholesterol concentrations in an epidemiological study in human subjects( Reference Ross, Bruce and Blondel-Lubrano 45 ). Owing to their similarities to humans in anatomy, body size, physiology, metabolism and omnivorous habits, pigs are better-suited for human metabolic studies when compared with rodents( Reference Miller and Ullrey 46 , Reference Neeb, Edwards and Alloosh 47 ). Although mostly descriptive, the results presented herein provide the first evidence that maternal dietary betaine supplementation during gestation causes complex and different changes in the hepatic expression of cholesterol metabolic genes in neonatal piglets, with the involvement of epigenetic modifications including DNA and histone methylation, as well as miRNA expression. These findings may help understand the role of maternal betaine supplementation in fetal programming of cholesterol metabolism in humans.
Obviously, the present study has some limitations. First, epigenetic regulation is only one of the possible mechanisms underlying the altered hepatic expression of cholesterol metabolic genes. The participation of other regulatory mechanisms, such as transcriptional factors and their interaction with epigenetic mechanisms, cannot be ruled out. Second, changes in hepatic cholesterol metabolism in neonates may persist to adulthood, causing long-term consequences in cholesterol homeostasis later in adult life. Follow-up studies are necessary to address the long-term effects on adult health and disease in response to the fetal programming of hepatic cholesterol metabolism caused by maternal betaine supplementation.
Supplementary material
To view supplementary material for this article, please visit http://dx.doi.org/10.1017/S0007114514002402
Acknowledgements
The present study was supported by the National Basic Research Program of China (2012CB124703), the Special Fund for Agro-Scientific Research in the Public Interest (201003011), the Fundamental Research Funds for the Central Universities (KYZ200913), the Priority Academic Program Development of Jiangsu Higher Education Institutions and the Innovation Project of Jiangsu Province Postgraduate Education (2013CXLX13_292).
We thank Dr Haoyu Liu (Uppsala University, Sweden) for critical comments on the manuscript.
The authors’ contributions were as follows: D. C. performed the experiments and measurements of serum biochemical parameters, analysed and interpreted the results, and drafted the manuscript; Y. J. and J. L. determined the serum hormone and amino acid levels; M. Y., S. S. and H. S. contributed to the experimental design; R. Z. contributed to the experimental concepts and design, provided scientific direction, analysed and interpreted the results, and finalised the manuscript. All authors read and approved the final manuscript.
There are no conflicts of interest to declare.









