Introduction
Wheat (Triticum aestivum L.) is an important food crop worldwide (Asseng et al., Reference Asseng, Guarin, Raman, Monje, Kiss, Despommier, Meggers and Gauthier2020). Drought is one of the most important abiotic stresses constraining wheat production (Gupta et al., Reference Gupta, Rico-Medina and Caño-Delgado2020). The annual crop yield reduction caused by drought exceeds the sum of other abiotic factors (Lesk et al., Reference Lesk, Rowhani and Ramankutty2016). Drought can lead to a sharp drop in production; it has been estimated that 42.0% of wheat cultivated are afflicted by drought in the world (Gupta et al., Reference Gupta, Rico-Medina and Caño-Delgado2020). Breeding wheat cultivars with strong drought tolerance is one of the effective ways to maintain a stable yield under drought conditions as global warming (Khadka et al., Reference Khadka, Earl, Raizada and Navabi2020). Seed germination is an initial period of wheat growth and development that is fragile to drought stress (Mickky and Aldesuquy, Reference Mickky and Aldesuquy2017). Germination of wheat directly affects the speed and quality of seedling emergence, which determines the number of seedlings and significantly influences grain yield. Therefore, drought tolerance of wheat during germination is very important to obtain sufficient seedlings and high yield.
Plant drought tolerance is a typical quantitative trait (Zhu, Reference Zhu2002). Quantitative trait loci (QTL) analysis is an effective strategy for dissecting QTL and has been successfully applied for gene mining in crops (Liu et al., Reference Liu, Sukumaran, Claverie, Sansaloni, Dreisigacker and Reynolds2019). Previous studies have reported on drought tolerance at the wheat germination stage using QTL mapping (Yuan et al., Reference Yuan, Li, Tian and Han2011; Czyczyło-Mysza et al., Reference Czyczyło-Mysza, Marcińska, Skrzypek, Cyganek, Juzoń and Karbarz2014; Nagel et al., Reference Nagel, Navakode, Scheibal, Baum, Nachit, Röder and Börner2014; Ashraf et al., Reference Ashraf, Shahzad, Karamat, Iqbal and Ali2015; Liu et al., Reference Liu, Zhang, Hu, Wang, Gao, Liang and Feng2017). Two QTL located on chromosomes 3B and 6A, related with coleoptile length (CL) in durum wheat under osmotic stress, explained 8.9 and 12.1% of the phenotypic variances, respectively (Nagel et al., Reference Nagel, Navakode, Scheibal, Baum, Nachit, Röder and Börner2014). Two loci Qgrd2C and Qgpd2C on wheat chromosome 5B affected germination rate (GR) and germination percentage under drought, accounting for 6.0–10.0% of the phenotypic variances (Ashraf et al., Reference Ashraf, Shahzad, Karamat, Iqbal and Ali2015). QTL for wheat drought tolerance coefficient (DTC) were found at seedling stage in an F8:9 recombinant inbred line (RIL) population and three QTL, QRLR-WL-1D, QCLR-WL-3D and QPHR-WL-7A, were identified on chromosomes 1D, 3D and 7A, for the longest root length (RL), a ratio of CL and seedling height, respectively, explaining 10.2–13.1% of the phenotypic variance (Liu et al., Reference Liu, Zhang, Hu, Wang, Gao, Liang and Feng2017).
With the swift developments in wheat genomics during the last two decades, single nucleotide polymorphism (SNP) arrays have become an important tool for gene mining and crop improvement (Rasheed et al., Reference Rasheed, Hao, Xia, Khan, Xu, Varshney and He2017; Rasheed and Xia, Reference Rasheed and Xia2019). A large number of SNPs were detected to be related with several traits using the wheat 9 K array during seed germination under polyethylene glycol (PEG6000) (Thabet et al., Reference Thabet, Moursi, Karam, Graner and Alqudah2018). Using the 660 K array, 18 QTL were detected for measured traits in 150 doubled haploid (DH) lines derived from the cross between Hanxuan 10 and Lumai 14 and QESNP-DS-R2 on chromosome 5D explained 29.0% of the phenotypic variance (Li et al., Reference Li, Mao, Wang, Chang, Reynolds and Jing2019).
The DTC was calculated following traits of the genotypes in environments with and without water restriction for evaluation and selection of drought-tolerant genotypes (Blum and Jordan, Reference Blum and Jordan1985; Li et al., Reference Li, Mao, Wang, Chang, Reynolds and Jing2019). Therefore, it is effective to map genes for drought tolerance using the DTC (Frova et al., Reference Frova, Krajewski, Di Fonzo, Villa and Sari-Gorla1999). QTL analysis of DTC-related traits in food crops has been performed (Li et al., Reference Li, Mei, Mei, Liu, Fu, Zhou and Tu2014, Reference Li, Mao, Wang, Chang, Reynolds and Jing2019; Guo et al., Reference Guo, Yang, Chang, Ma, Tu, Xiong, Jiang, Feng, Huang and Yang2018). However, studies based on the DTC at wheat germination are limited. The drought-responsive genes TaSNAC8-6A and TaEXPA2 were significantly associated with drought tolerance in wheat seedlings (Mao et al., Reference Mao, Li, Wang, Cheng, Li, Mei, Chen and Kang2020; Yang et al., Reference Yang, Zhang, An, Li, Chen, Zhao, Wu, Wang, Hao, Wang and Wang2020). DNA sequence polymorphism analysis and gene mapping were employed to develop functional markers TaCRT-D and TaP5CS, which are useful for the improvement of drought tolerance in wheat breeding (Wang et al., Reference Wang, Li, Mao and Jing2017; Yu et al., Reference Yu, Wei, Chang, Yang, Wu, Chen and Zhang2021).
Enhancing the diversity of wheat germplasm is one of the important goals in the International Maize and Wheat Improvement Center (CIMMYT) (Guzmán et al., Reference Guzmán, Autrique, Mondal, Huerta-Espino, Singh, Vargas, Crossa, Amaya and Peña2017). A large number of CIMMYT wheat cultivars have been successfully introduced into China in the past decades, which provide a quantity of breeding materials to overcome the bottleneck of simplification of germplasm resources in wheat breeding (Huang et al., Reference Huang, Wu, Wang, Mu, Xu, Zeng, Liu, Wang, Kang and Han2019).
The aims of this study were to (1) construct a high-density linkage map in the Berkut/Worrakatta RIL population using the wheat 50 K SNP array and (2) identify QTL for drought tolerance based on germination related traits using SNP-based genome-wide scanning.
Materials and methods
Plant materials
An F6 RIL population of 309 lines derived from a cross between CIMMYT spring wheat cultivars Berkut (pedigree: Irena/Babax//Pastor) and Worrakatta was used in the study. Berkut and Worrakatta showed moderate and high drought tolerance, and the comprehensive evaluation D-values of Berkut and Worrakatta were 0.17 and 0.57, respectively, to PEG6000 at the germination stage. All RILs and parents were grown in Manasi (44.18°N, 86.13°E) in Xinjiang province of China in 2018. Field trials were arranged in a randomized complete block design with three replications. Eighty seeds of each line were planted in 2 m rows with a spacing of 20 cm between rows. Field management was performed according to local practices. Seeds of parents and RILs were harvested for subsequent experiments.
Seed drought stress treatments
Trials with control and stress treatments were carried out to evaluate the germination related traits in the RILs and parents from August 2018 to January 2019. To exclude the influence of other organisms on the trials, wheat seeds were disinfected before the stress treatment. Six hundred seeds of each line with uniform size, full-grain and free of pest were selected and dipped into 70.0% ethanol for 1 min, washed in distilled water for five times and then soaked in 0.1% HgCl2 solution for 15 min, rinsed with distilled water for five times. One hundred seeds of each line were placed randomly and uniformly on single-layer filter papers on a germination dish (10 cm × 10 cm × 5 cm) with three replications for the PEG6000 treatment and three control treatments. Subsequently, 12 ml of 20.0% (W = −0.50 MPa) PEG6000 solution was added to the first three replications, while the same amount of deionized water was used for the three replications of control. The germination dishes were kept in a Percival intelligent light incubator (Model LT-36VL, www.percival-scientific.com) at 20 °C for an 18-h light/6-h dark photoperiod, with a light intensity of 150 μmol/m2 s and 60.0% of relative humidity for 7 d.
Trait measurements
The germination criterion was that the radicle length was equal to the seed length, or the germ length was larger than or equal to 1/2 of the seed length. Seven traits related to germination were evaluated. The germination potential (GP) was determined as GP = (n/N) × 100%, where n represents the number of germinated seeds on the 3rd day and N represents the number of total seeds. The GR was calculated by the formula GR = (n/N) × 100%, where n represents the number of germinated seeds on the 7th day and N represents the number of total seeds. Germination index (GI) = Σ (Gt/Dt), where Gt represents the number of germinated seeds on the t-th day and Dt represents the corresponding germination days. After the 7th day of germination, 10 germinated seedlings each replication were randomly selected, and the root number (RN), RL, shoot height (SH) and CL were measured with a ruler. The DTC was calculated using the equation: DTC = the measured values under PEG stress/the measured values of the control. The drought tolerance involving multiple traits was successfully judged by the comprehensive drought tolerance evaluation (D-value) based on DTC in the tested cultivars (Osipova et al., Reference Osipova, Permyakov, Permyakova, Rudikovskaya, Pomortsev, Verkhoturov and Pshenichnikova2020; Zou et al., Reference Zou, Hu, Li, He, Zhu and Zhou2020). The D-value was calculated by the formula of Zou et al. (Reference Zou, Hu, Li, He, Zhu and Zhou2020).
Genotyping and genetic map construction
The genomic DNA of parents and RILs was extracted from fresh leaves with a modified CTAB method (Saghai-Maroof et al., Reference Saghai-Maroof, Soliman, Jorgensen and Allard1984). All RILs and parents were genotyped using the wheat 50 K SNP array (http://www.capitalbiotech.com/).
Heterozygous loci were judged to be missing data, and SNPs with missing data more than 20.0% or allelic frequencies below 0.30 and over 0.70 were removed in subsequent analysis. High-quality SNPs retained were binned by the ‘BIN’ function in QTL IciMapping V4.1 (http://www.isbreeding.net/), and frame markers with minimum missing data were chosen; the groups of frame markers were sorted by the ‘Grouping’ function in JoinMap V4.0 with the logarithm of odds (LOD) thresholds ranging from 3 to 20 (Stam, Reference Stam1993). Genetic distances between markers were determined by the Kosambi mapping function (Kosambi, Reference Kosambi1944). The physical locations of markers in linkage groups were determined based on the wheat reference genome in the IWGSC RefSeq v2.0 database (https://urgi.versailles.inra.fr/blast_iwgsc/blast.php), and the maps were drawn in MapChart V2.3.2 (Voorrips, Reference Voorrips2002).
Data analysis and QTL mapping
The mean values of each RIL from three replicates were used for subsequent statistical analysis. Analysis of variance (ANOVA) was performed by the ‘AOV’ tool in QTL IciMapping V4.1 (Meng et al., Reference Meng, Li, Zhang and Wang2015). The broad-sense heritabilities (hB 2) were calculated following Nyquist and Baker (Reference Nyquist and Baker1991).
QTL was detected by the inclusive composite interval mapping (ICIM) method in the software QTL IciMapping V4.1 (http://www.isbreeding.net/) by setting the walking speed for genome scanning to 1.00 cM with P < 0.001. Significant LOD thresholds were determined using 1000 permutation tests with Type I Error at P < 0.05. QTL were named following the rules of International Rules of Genetic Nomenclature (https://wheat.pw.usda.gov/ggpages/wgc/98/Intro.htm), where ‘xjau’ represents ‘Xinjiang Agricultural University’.
Search for candidate genes
To predict candidate genes associated with QTL for drought tolerance related traits at germination in the Berkut/Worrakatta population, flanking sequences of the closest linked markers were used to blast the database of the Chinese Spring reference genome (IWGSC RefSeq v2.0, https://urgi.versailles.inra.fr/blast_iwgsc/blast.php), and their physical intervals on the reference genome sequences were obtained. Then, the annotation of genes was found in the physical intervals of the SNP markers in EnsemblePlants (http://plants.ensembl.org/index.html).
Results
Phenotypic evaluation
ANOVA exhibited significant differences in genotypes, treatments and genotype × treatment interactions (P < 0.01) for all investigated traits (GP, GR, GI, RL, SH and CL) except for RN under different treatments (online Supplementary Table S1). Broad-sense heritabilities (hB 2) ranged from 0.66 to 0.87. GP, GR and GI had higher hB 2 (above 0.85), indicating that the variations of the measured traits were mainly determined by genotypes. The average phenotypic values of all traits displayed transgressive segregation and continuous variation (online Supplementary Table S2). There were significant differences (P < 0.01) in the mean values of each trait among the RILs under different treatments. The coefficient of variation (CV) ranged from 10.0% for RN under the control treatment to 55.9% for GP under the PEG treatment. The measured traits (GP, GR, GI, RL, DTC_GR, DTC_GI and D-value) conformed to the normal distribution, showing that they were determined by multiple genes. There was a highly significant positive correlation (P < 0.01) among all traits (online Supplementary Tables S3 and S4). Among them, a high and positive correlation between GP and GI was observed (r = 0.91).
Linkage map construction
Twenty-eight linkage groups were constructed for the 21 wheat chromosomes, in which 15 chromosomes were corresponding to single linkage groups, while chromosomes 1A, 2A, 2D, 4A, 5A and 6D were divided into two or more linkage groups (Table 1). This genetic map contains 11,375 markers, which represented 1604 bins information and covered a total length of 2220.26 cM. The individual chromosomes range from 37.56 cM (6D) to 175.47 cM (5D), with an average length of 105.73 cM. The markers on linkage maps range from 214 (2D) to 1114 (5B), with 37.6%, 40.5% and 21.7% on A, B and D genomes, respectively. The average distance between bin markers was 1.38 cM.
Table 1. Summary of marker numbers and genetic distances of linkage groups in the Berkut/Worrakatta population

QTL mapping and clustering
Eighteen QTL for drought tolerance related traits at germination were identified in the Berkut/Worrakatta population (Table 2, Fig. 1). Among them, three for GP were mapped on chromosomes 4DS, 5DL and 6DS, one for GR on chromosome 5AS, two for GI on chromosomes 4DS and 5DL, one for RL on chromosome 1DL, four for SH on chromosomes 5AS, 5DL, 7AL and 7DS, two for GL on chromosomes 3DL and 5AS and five for D-value on chromosomes 1DL, 3DL, 5AS, 5BS and 5DL, respectively. The individual QTL explained 2.7–6.5% of the phenotypic variances.
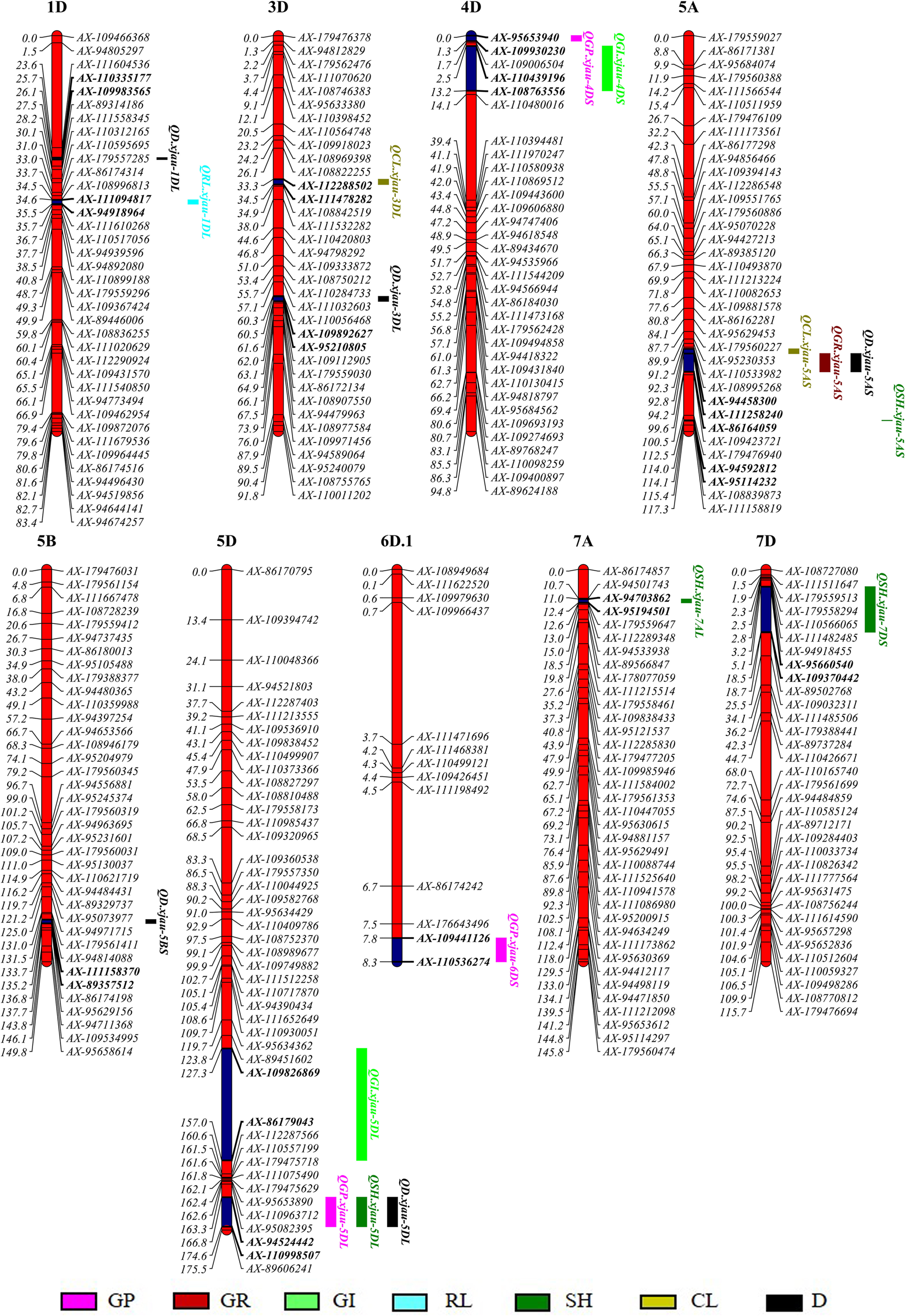
Fig. 1. QTL for drought tolerance related traits at germination stage on the Berkut/Worrakatta consensus map. GP, germination potential; GR, germination rate; GI, germination index; RL, root length; SH, shoot height; CL, coleoptile length; D-value, comprehensive drought tolerance evaluation.
Table 2. Positions and effects of drought tolerance related QTL detected at germination stage in the Berkut/Worrakatta RIL population

The positive alleles of QGP.xjau-4DS, QGI.xjau-4DS, QSH.xjau-7AL, QSH.xjau-7DS and QD.xjau-5BS were derived from Worrakatta, whereas those of QGP.xjau-5DL, QGP.xjau-6DS, QGR.xjau-5AS, QGI.xjau-5DL, QRL.xjau-1DL, QSH.xjau-5AS, QSH.xjau-5DL, QCL.xjau-3DL, QCL.xjau-5AS, QD.xjau-1DL, QD.xjau-3DL, QD.xjau-5AS and QD.xjau-5DL were from Berkut. Three QTL, QGR.xjau-5AS, QCL.xjau-5AS and QD.xjau-5AS, for GR, CL and D-value, respectively, at physical positions of 11.70–20.61 Mb between markers AX-111258240 and AX-94458300 on chromosome 5AS accounted for 3.4–4.8% of the phenotypic variances. The other three QTL, QGP.xjau-5DL, QSH.xjau-5DL and QD.xjau-5DL, for GP, SH and D-value, respectively, were flanked by markers AX-94524442 and AX-110998507 at 560.42–567.39 Mb on chromosome 5DL, accounting for 4.4–6.5% of the phenotypic variances. Hence, these two marker intervals are important pleiotropic loci. Six QTL were clustered in two intervals on chromosomes 5AS and 5DL, respectively (online Supplementary Table S5). One cluster (C5A) for GR, CL and D-value on chromosome 5AS was observed between SNPs AX-111258240 (11.70 Mb) and AX-94458300 (20.61 Mb), while the other (C5D) on chromosome 5DL (AX-94524442-AX-110998507) affected GP, SH and D-value, explaining 3.4–6.5% of the phenotypic variances. It is worth noting that all the favourable alleles in the two regions were derived from Berkut. Therefore, C5A and C5D stand for a ‘hot-spot’ with pleiotropic QTL for drought stress at germination.
Candidate genes
Five QTL for D-value were identified in physical intervals of less than 15.80 Mb. The QTL QD.xjau-5AS, QD.xjau-5BS and QD.xjau-5DL were detected in 7.40, 0.30 and 6.97 Mb regions, respectively (Table 2). A total of 198 high confidence genes were found in the physical intervals for these QTL (online Supplementary Table S6). According to the genome annotation information, three genes were targeted (Gene ID: TraesCS5A02G022100, TraesCS5B02G014200 and TraesCS5D02G563900), tentatively named as TaGATAs-5A, TaUbox-5B and TaGSTP-5D, encoding GATA transcription factor, RING/U-box superfamily protein and Glutathione S-transferase (GST), respectively (Table 3). To further identify candidate genes, wheat abiotic stresses transcriptional expression data were used (WheatOmics, http://202.194.139.32/expression/wheat.html). The results showed that TaGATAs-5A had the highest expression level under control, while the expression in tolerant variety Giza168 decreased with the increase of treatment time under PEG6000, and the drought stress was similar at the seedling stage (online Supplementary Table S7). Under PEG6000 stress, the expression level of TaUbox-5B reached highest after the treatment for 2 h in the tolerant variety Giza 168 and after 10–12 h in the sensitive variety Gemmiza 10. Under the treatment of PEG6000, the expression of TaUbox-5B can rapidly increase to respond on stress in the drought-resistant cultivar. The expression of TaUbox-5B was highest after 6 h of drought stress at seedling stage. The TaGSTP-5D expression is similar to TaUbox-5B in tolerant and sensitive varieties. They are likely to be candidate genes for regulating drought tolerance.
Table 3. Candidate genes corresponding to SNPs closely linked to the QTL

Discussion
Comparison of QTL in the present study with previous reports
Polyethylene glycol (PEG) as an osmotic regulator is widely used in the study of drought tolerance at seed germination in wheat (Duan et al., Reference Duan, Zhu, Li, Ding, Wang, Jiang and Zhou2017). Rapid germination, development of a long coleoptile and establishing a good root system are important requirements for wheat production in drought-prone areas (Heřmanská et al., Reference Heřmanská, Středa and Chloupek2015). GP, GR, GI, RN, RL, SH and CL are often used as indices for drought tolerance at the germination stage (Czyczyło-Mysza et al., Reference Czyczyło-Mysza, Marcińska, Skrzypek, Cyganek, Juzoń and Karbarz2014; Nagel et al., Reference Nagel, Navakode, Scheibal, Baum, Nachit, Röder and Börner2014; Ashraf et al., Reference Ashraf, Shahzad, Karamat, Iqbal and Ali2015). In the present study, 18 QTL were identified for drought tolerance related traits, including 15 new QTL for GP, GR, GI, CL and D-value.
QRL.xjau-1DL for RL on chromosome 1D explained 4.3% of the phenotypic variance, while QRLR-WL-1D for the DTC of the RL was located at 37.54 Mb between wpt-5503 and Xgpw7082.2 on chromosomes 1D at germination (Liu et al., Reference Liu, Zhang, Hu, Wang, Gao, Liang and Feng2017). These results suggest that QRL.xjau-1DL is a new QTL for RL. QSH.xjau-7AL for DTC of SH was detected on chromosome 7AL (607.61–722.93 Mb), in a similar position to QPHR-WL-7A for DTC of seedling height (Liu et al., Reference Liu, Zhang, Hu, Wang, Gao, Liang and Feng2017). Meanwhile, a significant association between marker M14627 on chromosome 7A and shoot biomass dry weight/m−2 was identified (670.75 Mb) under drought stress, and the gene TraesCS7A02G167900 was predicted (Mathew et al., Reference Mathew, Shimelis, Shayanowako, Laing and Chaplot2019), indicating that the two QTL are equal. QSH.xjau-7DS was found on chromosome 7DS (16.72–24.99 Mb), while a significant associated locus of shoot biomass dry weight/m2 was detected on chromosome 7D (99.95 Mb) under drought stress that is far away from QSH.xjau-7DS (Mathew et al., Reference Mathew, Shimelis, Shayanowako, Laing and Chaplot2019). Khalid et al. (Reference Khalid, Gul, Amir, Ali, Afzal, Quraishi, Ahmed and Rasheed2018) located QSL.nust-7D on the 295.00 cM of chromosome 7D under water-limited conditions, but there is a 286 cM distance from QSH.xjau-7DS.
Liu et al. (Reference Liu, Zhang, Hu, Wang, Gao, Liang and Feng2017) detected QCLR-WL-3D for the ratio of CL, which is at least 70 Mb far from QCL.xjau-3DL in this study. QCl3D-b flanking markers were located in 462.53–564.60 Mb on chromosome 3D (Yuan et al., Reference Yuan, Li, Tian and Han2011), which is close to QCL.xjau-3DL at 530.72–537.98 Mb. Therefore, this region can be a key interval for discovering coleoptile-related genes under drought stress. Five QTL for D-value were mapped on chromosomes 1DL, 3DL, 5AS, 5BS and 5DL; these are likely to be new loci, providing an important resource for the improvement of drought resistance in wheat breeding.
QTL clusters
In the present study, we found two QTL clusters (C5A and C5D) on chromosomes 5AS and 5DL, respectively. The physical position of C5A is similar to the chromosome region in Qaseem et al. (Reference Qaseem, Qureshi, Muqaddasi, Shaheen, Kousar and Röder2018), who reported a pleiotropic region on chromosome 5AS. The pleiotropic region at 11.70–20.61 Mb on chromosome 5AS was flanked by markers AX-111258240 and AX-94458300 associated with GR, CL and D-value under drought stress at germination, and also related to reduced grain yield under drought conditions. Under drought stress, the mobilization of reserves from the stem plays an important role in the supply of carbohydrates to grains at the stage of grain development (Blum, Reference Blum1996). Salem et al. (Reference Salem, Röder and Börner2007) found a major QTL (QSrm.ipk-5D) for stem reserve mobilization under drought stress, which is at least 150 cM far from C5D. In subtropical and arid areas, heat and drought are very severe problems at flowering and filling stages, which is the most fragile stage directly affecting grain yield (Ortiz et al., Reference Ortiz, Sayre, Govaerts, Gupta, Subbarao, Ban, Hodson, Dixon, Ortiz-Monasterio and Reynolds2008). The adverse effect of heat stress on wheat is that the duration of grain filling is shortened, which is mainly due to the decreased efficiency of the photosynthetic device itself and/or the decreased supply of photosynthetic assimilates caused by the loss of chlorophyll (Sharma et al., Reference Sharma, Torp, Rosenqvist, Ottosen and Andersen2017). A major and stable QTL for grain filling duration (QHgfd.iiwbr-5A) were identified for heat tolerance in wheat (Sharma et al., Reference Sharma, Torp, Rosenqvist, Ottosen and Andersen2017). Due to the lack of marker sequence information, the physical location of the QTL cannot be determined. Thus, it is impossible to judge whether it is closely linked to the C5A found in this study. Root architecture and system are the key structure under drought conditions (Wasaya et al., Reference Wasaya, Zhang, Fang and Yan2018). Salarpour et al. (Reference Salarpour, Pakniyat, Abdolshahi, Heidari, Razi and Afzali2020) reported a stable QTL on chromosome 5A (35.63–41.42 Mb; Excalibur_rep_c68688_103 and Kukri_c14944_771) associated with RN under drought stress, which is closely linked to AX-111258240-AX-94458300 in the present study. The marker wsnp_BE399966A_Ta_2_3 that was significantly associated with tiller number on chromosome 5A (9.65 Mb) under drought stress (Abou-Elwafa and Shehzad, Reference Abou-Elwafa and Shehzad2021) is tightly linked to the C5A QTL cluster for GR, CL and D-value in the present study. The above QTL clusters provide important information for tolerance to abiotic stresses in wheat. The adaptability of wheat to various environmental conditions mainly depends on the allelic diversity of genes controlling vernalization requirement (Vrn-1) (Royo et al., Reference Royo, Dreisigacker, Soriano, Lopes, Ammar and Villegas2020). Vrn-1 is located on the long arm of the homologous group of chromosome 5 in wheat, where Vrn-D1 and C5D are at least 90 Mb apart (Yan et al., Reference Yan, Loukoianov, Tranquilli, Helguera, Fahima and Dubcovsky2003). Thus, chromosome 5A and 5D possibly involves many resistance genes that can be used as candidates for genetic analysis.
Candidate gene prediction
Based on the reference wheat genome and genetic map, the potential genes for the identified QTL involved in the drought tolerance at the germination stage in this study were postulated. Transcription factor TaGATAs-5A may be the causal gene of QD.xjau-5AS. In plants, transcription factors are involved in growth, development, abiotic and biotic stresses (Joshi et al., Reference Joshi, Wani, Singh, Bohra, Dar, Lone, Pareek and Singla-Pareek2016). GATA factor is a kind of transcriptional regulator, which widely expresses in fungi, animals and plants (Lowry and Atchley, Reference Lowry and Atchley2000). The DNA binding domain of GATA factor consists of type IV zinc finger of CX2CX17-20CX2C and a highly basic region (Ko and Engel, Reference Ko and Engel1993). Stress-related signals stimulate the binding of the zinc-finger protein gene with the corresponding cis-acting elements. It promotes the activation of RNA polymerase II transcriptional complex and turns on the transcriptional expression of specific genes. In rice, OsGATA8 is a homologous gene of TaGATAs-5A, and it is the main regulator of the plant stress response and regulates the expression of downstream stress resistance and ROS-scavenging enzymes genes and participates in the interaction of different cellular mechanisms under stress (Nutan et al., Reference Nutan, Singla-Pareek and Pareek2020).
The candidate gene TaUbox-5B was detected in QD.xjau-5BS region. The accumulation of abnormal proteins, such as unfolded or misfolded proteins, may damage cell function and eventually lead to cell death (Hatakeyama and Nakayama, Reference Hatakeyama and Nakayama2003). Key regulatory components in the cellular process include protein degradation (Dunlap, Reference Dunlap1999). Intracellular proteolysis is mainly mediated by ubiquitin-26S-proteasome system 3 (Ciechanover, Reference Ciechanover1998). The last step connects to the target protein and covalently binds to the substrate protein through ubiquitin ligase (Azevedo et al., Reference Azevedo, Santos-Rosa and Shirasu2001). U-box proteins belongs to the ubiquitin ligases family (Hatakeyama and Nakayama, Reference Hatakeyama and Nakayama2003). Previous studies showed that four U-Box negatively regulated abscisic acid-mediated pathway under drought stress in Arabidopsis (Seo et al., Reference Seo, Ryu, Jammes, Hwang, Turek, Kang, Kwak and Kim2012).
TaGSTP-5D was predicted as a candidate gene for QD.xjau-5DL. Reactive oxygen species (ROS) are produced by molecular oxygen excitation or incomplete reduction, that are harmful by-products of aerobic organisms (Apel and Hirt, Reference Apel and Hirt2004; Miller et al., Reference Miller, Suzuki, Ciftci-Yilmaz and Mittler2010). Plant cells produce excessive ROS under stress, which is highly reactive and toxic to proteins, lipids and nucleic acids, resulting in cell damage and death (Gill and Tuteja, Reference Gill and Tuteja2010). GST is a ROS-scavenging enzyme, that uses thioredoxin (TRX) as a nucleophilic to reduce organic hydroperoxides through an ascorbic pathway (Meyer et al., Reference Meyer, Belin, Delorme-Hinoux, Reichheld and Riondet2012). Fine mapping, haplotype analysis and functional confirmation of the target candidates of the QTL for drought tolerance will be focused on in our future studies.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S1479262121000551
Acknowledgements
This study was supported by the Foundation of Xinjiang Uygur Autonomous Regional Educational Committee (XJEDU2020I010), National Natural Science Foundation of China (31461143021) and a grant from the Xinjiang Uygur Autonomous Regional ‘Tianshan Xuesong’ project (2018XS04).
Author contributions
Hw-G, YR and Jx-Z designed and conceived the experiments, and YR performed all the experiments and wrote the manuscript. Jd-L assisted in analysing the data. Hw-G, Xc-X, and SD provided direction for the study and corrections to the manuscript. All authors read and approved the manuscript.
Conflict of interest
The authors declare no competing interests.






