Introduction
Measles is a highly contagious and serious childhood disease caused by the measles virus (MV). MV belongs to the paramyxovirus the paramyxovirus family, of the genus morbillivirus, is a single-stranded RNA virus and is an enveloped virus transmitted by direct contact or air. Then, the virus settles in mucous membranes and spreads to the body. Over the course of the four days before the rash begins and during the first 4 days following the onset of the rash, the risk of infection from person-to-person is high [1, Reference Murray2]. Although a safe and cost-efficient vaccine is available, measles has been a leading cause of death among children. In 2014, approximately 115 000 children under the age of five lost their lives due to measles [1]. Even in countries where measles has been eliminated to a large extent, imported or import-related cases received from other countries continue to be an important source of infection [Reference Murray2]. Timely measles surveillance is critical to disease control. Identifying and confirming suspected measles cases through surveillance allows for early detection of outbreaks, analysis of on-going transmission to mount more effective vaccination measures and estimation of the underlying true measles incidence based on the patterns in reported data. The genomic sequence analysis of the MV can help track its spread and monitor progress towards elimination. A measles–rubella laboratory surveillance network was established in Turkey in 2002 and has begun to provide services with seven sub-national laboratories and one national laboratory. Neighbouring provinces have been defined to those sub-national laboratories, and thus, suspected measles cases are detected in 81 provinces of the country in a laboratory network that includes sub-national laboratories and national laboratory. Because high vaccination rates have not been achieved among the World Health Organization-Europe Region, including Turkey, virus circulation on the continent of Europe could not have been prevented, and since 2005, various outbreaks have occurred. Consistent with increased people movement and the location of Turkey, imported cases come from abroad, and unvaccinated people or people with deficient vaccination may be affected by those cases and may become ill [3–Reference Muscat6]. The purpose of this study was to evaluate the laboratory findings of confirmed measles cases in the last nine years, to assess the epidemiological data of the cases, to determine the molecular genotyping studies and to emphasise the importance of laboratory-based surveillance of measles.
Materials and methods
Patients and sample selection
The measles/rubella monitoring network, established in 2002 and successfully implemented, enables the early detection of a measles outbreak in Turkey. Regarding patient and sample selection, case definitions have been conducted according to the measles and rubella infection laboratory diagnosis manual of the WHO and the Measles, Rubella and Congenital Rubella Syndrome Surveillance Permanent Circular, which was published by our Ministry of Health in 2010. Surveillance activities have been conducted accordingly [7, 8].
Measles case definition
Clinical criteria: Fever (>38 °C) and maculopapular rash and cough or coryza or conjunctivitis. Clinical criteria were present in all cases.
Laboratory criteria: Measles IgM antibody detection or measles viral RNA detection by RT–PCR.
For all suspected cases, the case investigation and laboratory request forms were completed. Serum, urine and nasopharyngeal swab samples were collected from patients who had attended health institutions in Turkey and had potential clinical symptoms (at least fever and maculopapular rash). Serum samples, taken from suspected measles cases were detected in 81 provinces in Turkey and were sent to eight laboratories in total, including even sub-national laboratories and one national laboratory, that are accredited by the WHO. Each laboratory tested the serum samples received from neighbouring provinces and provided a serum measles IgM result on the same day. Serum samples were taken within the first 28 days after the onset of the rash. Nasopharyngeal swab samples taken during the first 4 days of rash and urine samples taken during the first 9 days were also accepted. Nasopharyngeal swab samples and urine samples from patients who were measles IgM positive were sent to the National Laboratory for molecular tests. [7].
Serological and molecular diagnosis of MV
A total of 62 880 serum samples were tested for the presence of measles IgM with the manual microELISA method. A total of 3 different kits were used for this purpose over the years: Enzygnost®Anti-Measles Virus/IgM (Siemens, Marburg, Germany), Euroimmun Anti-Measles virus NP ELISA IgM (Luebeck, Germany) and Microimmun Measles IgM Capture EIA (Hounslow, UK). In addition, 688 urine or nasopharyngeal samples were tested. In the urine and/or nasopharyngeal samples of patients, viral nucleic acid extraction and isolation processes were performed. Extraction and isolation procedures were conducted according to the manufacturer's instructions with a Qiagen EZ1 Virus Mini Kit v2.0 (Qiagen, Hilden, Germany). The extraction product was researched in terms of MV RNA using a Real-Time Multiplex commercial kit (Fast Track Diagnostics, Luxembourg) and ABI 7500 Real-Time PCR device (Applied Biosystems, USA). In the present samples, by performing sequence analyses, the genotype was determined according to the sequence result.
Genotyping
Amplification of nucleocapsid (N) gene
The carboxyl terminus region of the N gene was sequenced for MV genotyping. Viral RNA was extracted using a viral RNAeasy Kit (Qiagen, Hilden, Germany). A One-Step Rt-PCR Master Kit (Qiagen; USA) was used to amplify a 634-bp C-terminal region of the N gene using the following primers: forward MeV214 (5′-TAACAATGATGGAGGGTAGG-3′) and reverse MeV216 (5′-TGGAGCTATGCCATGGGAGT-3′) [Reference Rota9]. In the single-phase test, cDNA synthesis was applied using the following amplification conditions: denaturation at 50°C for 30 min and at 95 °C for 15 min, 40 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, elongation at 72 °C for 1 min and elongation at 72 °C for 10 min. The PCR products were viewed using a 2% agarose gel containing 0.5 µg/ml ethidium bromide staining [Reference Maiwald10].
Sequence analysis
The PCR products were purified using a Beckman Coulter Agencourt AMPure XP kit (Beckman Coulter, Beverly, Mass., USA). Sequencing reactions were established and consisted of 3.5–5 ml of purified amplicon, 5 pmol primer and 4 ml of the dye terminator cycle sequencing Quick Start Kit (Beckman Coulter). The sequence reaction condition was the following: first denaturation at 94 °C for 3 min, 30 cycles of denaturation at 96 °C for 20 s, annealing at 55 °C for 30 s, elongation at 60 °C for 4 min. The sequence products were purified using a Dye-Terminator Removal Kit (Agencourt Cleanseq; Beckman Coulter). DNA capillary gel electrophoresis was conducted with the Beckman Coulter Ceq8000 (Beckman Coulter, Fullerton, CA, USA) system. Electrophoresis conditions were applied as 5 s injection times, 2 kV voltage, 4 kV electrophoresis voltage, 110 min duration, 50 °C capillary heat, 90 °C denaturation heat. The obtained sequences were analysed using National Center for Biotechnology Information (NCBI) GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and Clustal W interface (htttp:// www.ebi.ac.uk/clustal). Then, the sequences were entered into the Measles Nucleotide Surveillance Program (MeaNS) (http://www.hpa-bioinformatics.org.uk/Measles) data bank as country data. The MV Schwarz strain was used as a positive control in this analysis.
Results
Epidemiological findings
Since 2002, the coverage rate of the MCV-1 (measles-containing vaccine first dose) has been above 95%. MCV-2 (measles-containing vaccine second dose) was introduced to the immunisation schedule in 1998. Due to intensive supplementary immunisation activities and high routine immunisation coverage, especially during the last 7 years, a dramatic decrease in the number of measles cases has been recorded. The measles immunisation coverage rate in 1990 was 67% and reached 97% in 2015 (Fig. 1).
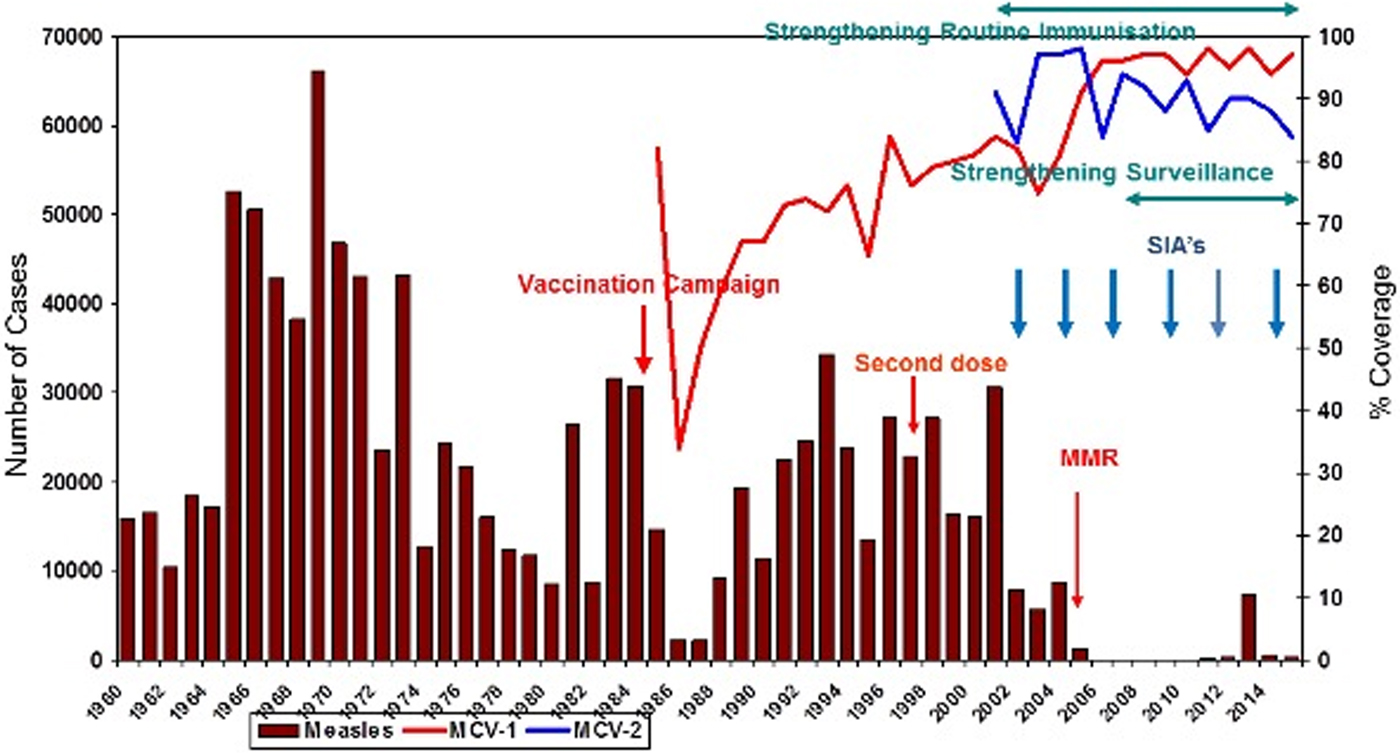
Fig. 1. Impact of measles elimination strategies in Turkey (1960–2015). MMR, Measles–Mumps–Rubella vaccination; MCV-1, measles-containing vaccine first dose; MCV-2, measles-containing vaccine second dose; SIA, supplementary immunisation activity.
In 2007, approximately 89% of all measles cases were from Turkey, 10% were from Syria and 1% were from other countries; 30% of cases occurred in children <1 year old, 25% in to those aged 1–4 years, 18% in patients aged 5–9 years and 27% in people over the age of 10 years old.
Laboratory findings
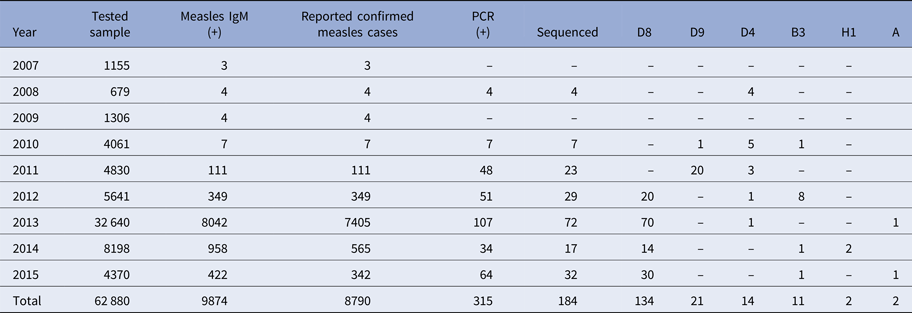
From 2007 to 2015, a total of 62 880 serum samples with a measles clinical diagnosis were sent to our national and sub-national laboratories and were tested for measles IgM, from which 9874 were positive for measles IgM. Of these samples, 8790 were accepted as measles cases (Fig. 2). The remaining IgM-positive findings were positives due to vaccination or false positives depending on other childhood rash diseases. Situations in which measles are detected as IgM positive but not measles cases can be listed as follows. Sequencing results measles genotype-A determination, positivity with rubella IgM due to MMR (measles, mumps, rubella) vaccination, parvovirus B19, enterovirus, HHV-6, HHV-7 positive and concurrent measles PCR negative detection by multiplex PCR, detection of rubella PCR positivity by reverse transcriptase PCR, detection of IgM positives by serologic tests for rash disease agents other than measles (parvovirus B19, varicella-zoster virus) and measles IgG positivity with measles IgG high avidity detection. In these cases, the clinical findings of the patient were observed and in connection with the epidemiology unit, the patient was removed from the measles classification. The vast majority of this group has been vaccinated due to the outbreak, resulting in positives due to the resulting antibody response. Of all samples tested, 64% were collected during the first 2 days from the onset of the rash and 36% were collected during the third and the following day (Fig. 3). Molecular tests could be performed on 688 measles IgM-positive cases where we received nasopharyngeal swab or urine specimen PCR positivity was detected in 315 (45.8%) samples. PCR-positive samples were subjected to DNA sequencing, and the sequence data of 184 samples were acquired (Table 1). The sequence data of 184 samples are shown in Figure 4 and include genotype D8 (n = 134), D9 (n = 21), D4 (n = 14), B2 (n = 11), H1 (n = 2) and A strain Schwarz (n = 2). In Turkey, the MV Schwarz strain has been used for vaccination. A total of 688 urine or nasopharyngeal/pharyngeal swab samples were tested by PCR and 315 samples were positive for measles PCR. PCR positivity was detectable only in 315 of 688 (45.8%) samples for which urine or swab samples could be obtained with serum. From 310 PCR-positive samples received during the first 5 days from onset of the rash, five patients were also PCR positive in urine, although the samples were collected between the 6th and 9th days.
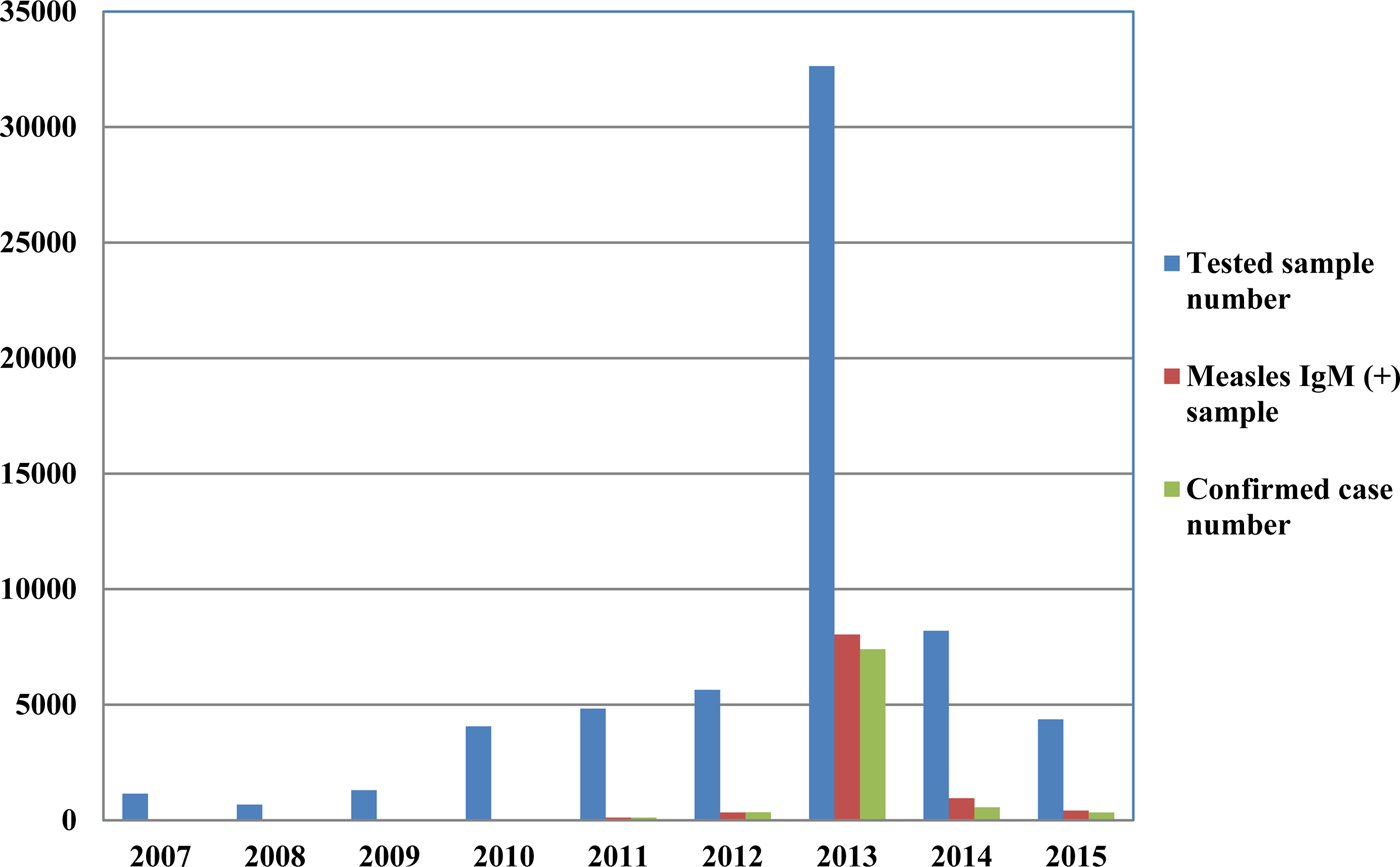
Fig. 2. Measles laboratory tests and case data between 2007 and 2015.
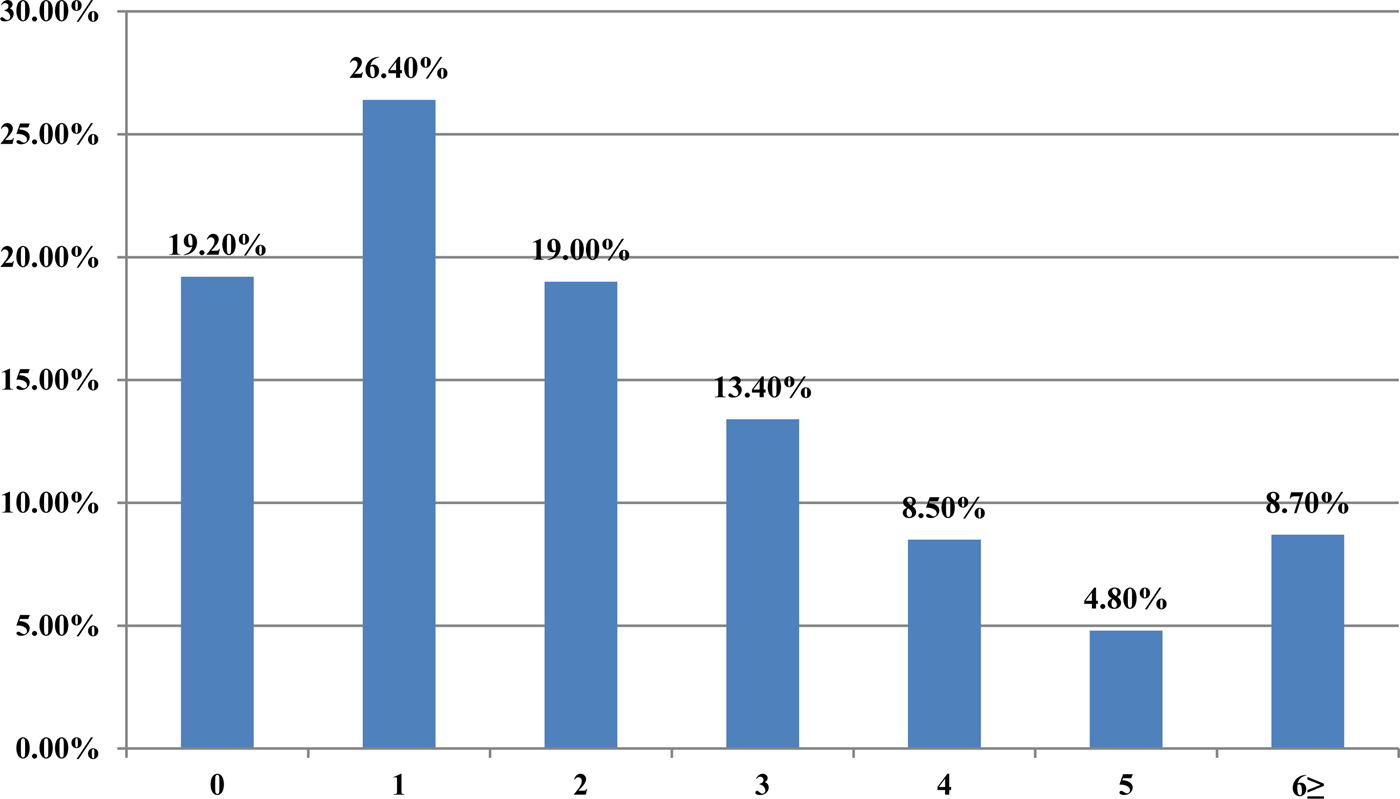
Fig. 3. The time between the onsets of rash-sampling time (days).
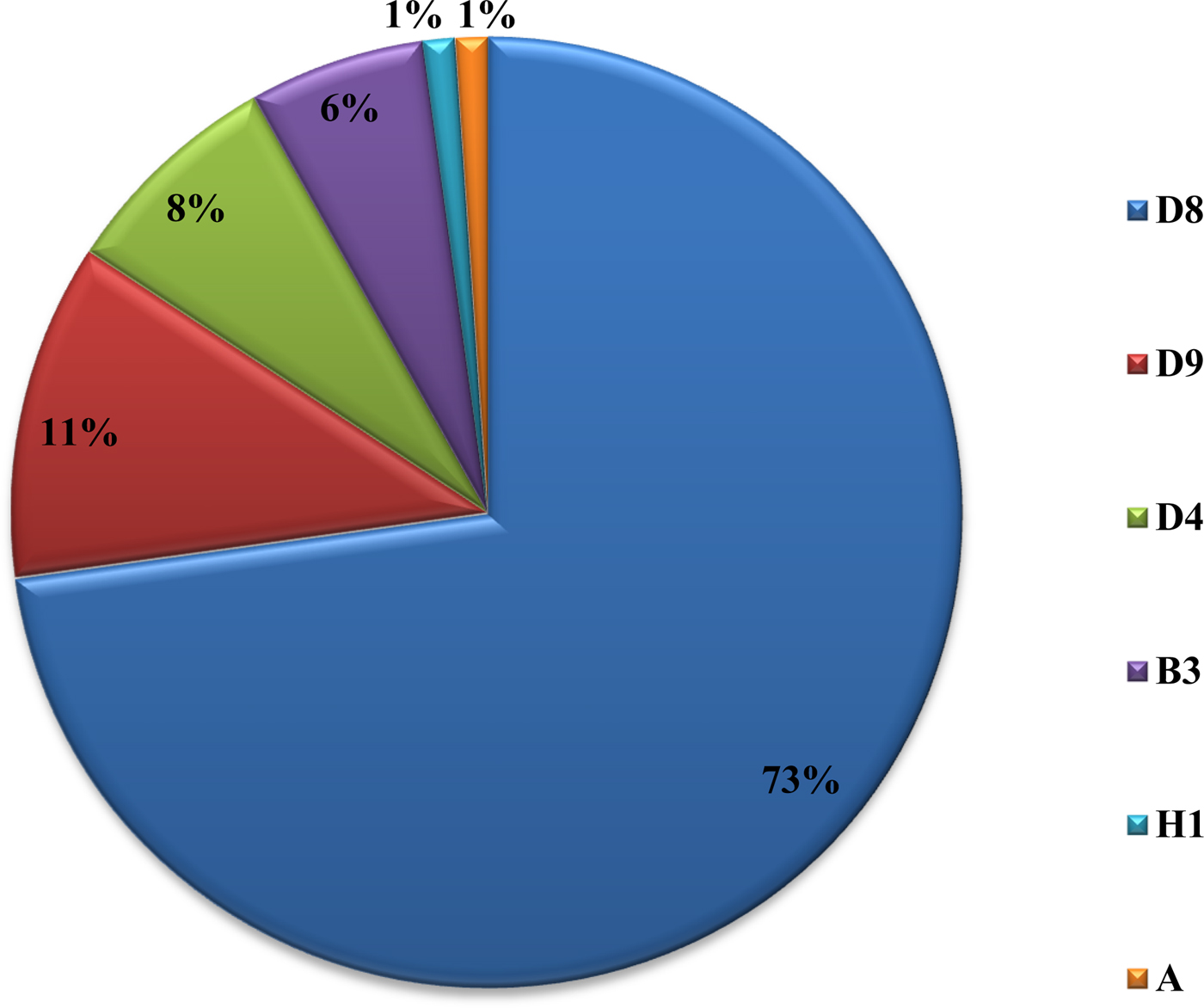
Fig. 4. Genotype distribution of MVs detected in Turkey between 2008 and 2015.
Table 1. The distribution of measles strains in circulation in our country by year
Discussion
A laboratory-based surveillance system is very important so that cases can be confirmed by a laboratory and their final diagnoses can be made. As there are clinical symptoms similar to childhood exanthem diseases in measles, the number of potential or suspected cases was high, especially during outbreaks. In our study, when the outbreak peaked in 2013, 32 640 patient samples with a measles clinical diagnosis were tested; however, measles IgM positivity was detected in 8042 (24.6%) samples. When clinically diagnosed, the number of measles patients was approximately four times the confirmed number of cases. In 2007, Kurugol et al. determined that only two of 34 (6%) patients meeting the measles case definition had measles IgM positivity [Reference Kurugol11]. Studies have indicated that the detection of viral RNA together with serum-specific measles IgM are supportive in the diagnosis. In the studies by Binnendijk et al. in 2003, 93% of patients with clinical symptoms tested positive for MV viral RNA, which was detected in an oropharyngeal swab [Reference Binnendijk12]. In our study, measles PCR positive was detected at 315 of urine and swab specimens obtained from 688 patients who were IgM positive and PCR negative was found in the remaining samples. We think that several reasons need to be investigated for this finding, for example, decrease or loss of viral load at the time of antibody response, inappropriate intake of swab specimens, factors related to the test method may play a role. Because serologic tests are cheap, easily applied, easily sampled, have high sensitivity and specificity, measles can be easily used to confirm the diagnosis in clinically diagnosed patients. However, PCR testing can also be used as a diagnostic test from nasopharyngeal swab or urine sample at the onset of the disease, especially in first 2 days when the antibodies are not at detectable levels and to help determine advanced genotypic analyses and molecular epidemiological studies. In our study, approximately 66% of the sera samples were collected in the first two days. The use of molecular tests together with serological tests in the cases collected in the first 2 days increases the sensitivity and specificity of measles. However, it is easier and more convenient to take a serum sample from patients with measles clinically diagnosed, while it may be difficult to obtain a nasopharyngeal swab or urine specimen [7]. Virologic surveillance is an important component of measles management for the elimination of the virus, which has become endemic in the country. It is also necessary to determine local genotypes and to conduct a genetic analysis of wild-type viruses and to detect imported cases [Reference Vainio13, Reference Bellini14]. A study conducted by Kalaycioglu et al. showed that genotype D9 viruses were detected in cases in 2011, and the identified viruses were similar to the D9 genotype detected in Russia, Malaysia, Japan and the UK [Reference Kalaycioglu15]. From 2012–2015, the D8 genotype viruses were considered to be spread from the Romanian buskers to Istanbul and became dominant until 2015 when the D8 outbreak became endemic. Sequence analyses of D8 genotype viruses detected in Turkey in 2012 and 2013 showed that these viruses were identical to the D8-Frankfurt-Main strain, which was detected in several countries during outbreaks in Europe [Reference Kalaycioglu16]. Two H1 genotypes were detected in two cases in 2014, and they were related to each other epidemiologically. Although the H1 genotype is endemic in western Pacific region countries, imported cases related to travelling may be experienced [Reference Xu17–Reference Sosa19]. Other viral agents, causing a similar clinical picture and differential diagnosis, have also been studied. Rubella IgM testing in addition to measles IgM was performed in received samples. Whereas measles PCR testing is performed, parvovirus B19, HHV-6,7 and enteroviruses have also been studied using multiplex PCR kits. Determining exanthema disease agents other than measles is important for determining the diagnosis of cases and monitoring contacts. When an epidemic occurs in a country, large-scale vaccination studies are carried out, and even though measles IgM positivity is detected serologically, clinical findings may be due to other rash diseases. In this case, the virology laboratory has a lot of work to determine the correct effect and give a result [Reference Ramdass20, Reference Scott21].
Conclusion
D9 outbreak was short and confined to 2010–2011, it may be from Russia. D4 was present from 2008 and continues to appear. B3 was also after 2010 and most in 2012. The D8 is a major problem and it appeared in 2012. It affects other parts of Europe too. The diagnosis of measles should be made by examining the clinical findings of the cases and at least the anti-measles IgM positivity of laboratory tests. The detection of measles RNA is important both for confirming the measles diagnosis and for genotyping the current virus to shed light on epidemiological molecular investigations. Laboratory-based measles surveillance is an important part of the measles elimination strategy. To control an outbreak, it is important to resolve under- or poor-vaccination coverage and monitor the disease with efficient epidemiologic and laboratory-based active surveillance systems, immediately test samples taken from patients and report the results as well as immediately reach the original case and contacts. Molecular tests must be performed together with serologic tests. For cases in which serologic tests are not positive yet, molecular tests can be used for diagnosis. The confirmation of potential measles cases by laboratory testing has an important role in controlling the outbreak by allowing direct isolation of the case and immediate vaccination of the contacts. Vital parameters in controlling the outbreak at the national and international levels include conducting a genotypic analysis of the virus in positive samples as well as an infection analysis with its epidemiological connections and taking planning measures based on the information.
Acknowledgements
We thank all field and laboratory employees involved in measles surveillance.








