Introduction
Bees (Hymenoptera: Apoidea) are well known for their role as pollinators, and have become the focus of much recent attention as populations are undergoing declines globally (e.g., Committee on the Status of Pollinators in North America 2007; Vamosi et al. Reference Vamosi, Gong, Adamowicz and Packer2016). Bees are a diverse taxon with estimates of over 20 000 species worldwide (Michener Reference Michener2007; Ascher and Pickering Reference Ascher and Pickering2017), with new species being described regularly, including 22 in Canada since 2010 (Gibbs Reference Gibbs2010; Rehan and Sheffield Reference Rehan and Sheffield2011; Gibbs et al. Reference Gibbs, Packer, Dumesh and Danforth2013; Williams et al. Reference Williams, Cannings and Sheffield2016). Brown and Paxton (Reference Brown and Paxton2009) stressed that alpha taxonomy, in particular lack of taxonomic expertise and resources, is one of the major factors limiting ability to conserve bees, since being able to accurately identify species and link them to specific habitats, crops or other plants they pollinate and/or the communities they service is critical for conserving them. However, taxonomic and faunistic knowledge of Canadian bees is incomplete for various reasons (see Sheffield et al. Reference Sheffield, Hebert, Kevan and Packer2009, Reference Sheffield, Frier and Dumesh2014; Droege et al. Reference Droege, Rightmyer, Sheffield and Brady2010), including the large number of poorly studied taxa, and the large size of the country resulting in many areas being poorly sampled. One challenge is underfunding of taxonomic training and research in general, including in Canada (Packer et al. Reference Packer, Sheffield, Gibbs, de Silva, Best and Ascher2009b), potentially contributing to slow rates of species discovery and taxonomic revisions (i.e., keys, descriptions and other tools for the identification of all species within a given taxon, including those already described) (see Carvalho et al. Reference Carvalho, Ebach, Williams, Nihei, Trefaut Rodrigues and Grant2014). This taxonomic impediment ultimately affects many ecological studies and conservation strategies for bees (e.g., Sheffield et al. Reference Sheffield, Richardson, Cannings, Ngo, Heron and Williams2016), though is even more of an issue for other taxa (e.g., Maurer Reference Maurer2000) that have not received the same level of attention.
Danks (Reference Danks1979) reviewed the diversity of terrestrial arthropods in Canada as an early initiative of the Biological Survey of Canada, this work considered one of the many outstanding achievements of the Biological Survey of Canada (Danks Reference Danks2016), this issue celebrating its 40th anniversary. That report (Danks Reference Danks1979) showed that the arthropod fauna was poorly documented, and pointed out that the 29 975 reported insect species likely represented only about half of the total expected for Canada. Since then, taxonomic advances (including new molecular taxonomy tools) have improved the knowledge base for many groups. For example, Hebert et al. (Reference Hebert, Ratnasingham, Zakharov, Telfer, Levesque-Beaudin and Milton2016) used DNA barcoding to estimate more than 94 000 insect species in Canada, about a 58% increase over Danks (Reference Danks1979) estimate, with Diptera and Hymenoptera (especially Parasitica) being “unexpectedly diverse” and in general poorly known.
In the Hymenoptera superfamily Apoidea, which includes the bees (Apiformes), ~800 species are currently known from Canada (Sheffield et al. Reference Sheffield, Frier and Dumesh2014), an increase of about 50 species since the estimates in Danks (Reference Danks1979; 746 species, though 231 more were predicted). No national checklist of bees for Canada has ever been completed, though regional lists for Nova Scotia (Sheffield et al. Reference Sheffield, Kevan, Smith, Rigby and Rogers2003), Newfoundland (Hicks Reference Hicks2009), and the Prairies Ecozone (Sheffield et al. Reference Sheffield, Frier and Dumesh2014) have. The bee fauna of Nova Scotia was also the first to be subjected to an extensive DNA barcoding effort (Sheffield et al. Reference Sheffield, Hebert, Kevan and Packer2009), starting in the early 2000s with ongoing diversity assessments (i.e., Sheffield et al. Reference Sheffield, Kevan, Smith, Rigby and Rogers2003, Reference Sheffield, Kevan, Westby and Smith2008).
Bee identification is well known to be challenging for several speciose genera in North America (see Packer et al. Reference Packer, Grixti, Roughley and Hanner2009a; Droege et al. Reference Droege, Rightmyer, Sheffield and Brady2010; Gibbs Reference Gibbs2010) due to lack of keys and taxonomic expertise for some taxa. Historically, even the greatest bee taxonomists of their time had trouble distinguishing species – T.D.A. Cockerell (1866–1948) described more bee species than any other person (with 3132 still considered valid today; Ascher and Pickering Reference Ascher and Pickering2017), but in several cases he (alone or as a coauthor) described the same species multiple times (sometimes in a single manuscript) that today are considered one. Early taxonomists (and likely many today) had limited ability to accurately link specimens to geography, associate sexes, etc., so often described specimens with slight variations in size and/or colour as new species or subspecies (or varieties). Allometric variation among castes in some eusocial Halictidae species (Gibbs Reference Gibbs2010), or linked to body size in Ceratina Latreille (Apidae) (Rehan and Sheffield Reference Rehan and Sheffield2011), has also likely contributed to the taxonomic difficulties observed in some bee taxa. Since then, taxonomists have seen the advent of high resolution microscopes, computers and extensive collection databases (e.g., Meier and Dikow Reference Meier and Dikow2004), wide and mostly free availability of taxonomic literature and other taxonomic resources (e.g., Biodiversity Heritage Library, university journal databases), and easier exchange of material for study within and among countries, including data sharing (including actual specimens and/or high-quality photographs, and specimen data). Incorporation of high-quality images into web-based interactive taxonomic keys (e.g., the mandate of the Canadian Journal of Arthropod Identification) has also facilitated accurate species level identification for the taxa that have received treatment. Despite these tools, many Canadian bee taxa are in need of revision. The challenge for today’s generation of taxonomists will be overcoming challenges to train the next generation of taxonomists to understand the range of software and procedures to construct and test phylogenies (morphological and molecular), species concepts, predictive models for distribution, and find employment to do so as a career. Ongoing work on bee taxonomy in Canada (primarily at York University, Toronto, Ontario, Canada) will be aided by the recent (since 2009) hiring of three additional full-time bee systematists in collection-based institutions (Royal Saskatchewan Museum, University of Manitoba, Canadian National Collection of Insects, Arachnids, and Nematodes), which has already resulted in a number of published works on the taxonomy of Canadian bees (Gibbs Reference Gibbs2010; Rehan and Sheffield Reference Rehan and Sheffield2011; Sheffield et al. Reference Sheffield, Ratti, Packer and Griswold2011b; Dumesh and Sheffield Reference Dumesh and Sheffield2012; Onuferko Reference Onuferko2017).
One important change in taxonomic research since the inception of the Biological Survey of Canada 40 years ago has been the development of molecular taxonomy tools. Genetic-based identification systems such as DNA barcoding (as per Hebert et al. Reference Hebert, Cywinska, Ball and deWaard2003a, Reference Hebert, Ratnasingham and de Waard2003b) have emerged as powerful and cost-effective tools for accurate identification of biota and assessing and understanding the extent of diversity in groups that have proven difficult using classical taxonomic techniques (Köhler Reference Köhler2007; Packer et al. Reference Packer, Grixti, Roughley and Hanner2009a). For North America, and Canada in particular, several DNA barcoding projects have been undertaken for pollinators (e.g., Sheffield et al. Reference Sheffield, Hebert, Kevan and Packer2009; Hebert and Landry Reference Hebert and Landry2010; Hebert and Humble Reference Hebert and Humble2011; Zahiri et al. Reference Zahiri, Lafontaine, Schmidt, Zakharov and Hebert2014, Reference Zahiri, Lafontaine, Schmidt, Zakharov and Hebert2017), and many other taxa. These DNA barcode libraries can assist traditional morphological taxonomy by allowing identification of life stages for which keys are not available and/or taxonomy is more difficult (e.g., Slowik and Blagoev Reference Slowik and Blagoev2012), associating sexes (e.g., Sheffield et al. Reference Sheffield, Ratti, Packer and Griswold2011b), and in studies assessing biological diversity within regional (e.g., Sheffield et al. Reference Sheffield, Hebert, Kevan and Packer2009) and poorly studied habitat-specific biotas (e.g., Smith et al. Reference Smith, Fisher and Hebert2005, Reference Smith, Fernandez-Triana, Roughley and Hebert2009; Stahlhut et al. Reference Stahlhut, Fernández-Triana, Adamowicz, Buck, Goulet and Hebert2013). DNA barcode sequences (i.e., cytochrome c oxidase subunit I (COI)) and other genetic sequences themselves also contribute to molecular systematics; combining the DNA barcode gene with a single nuclear gene has proven useful for accurate, node-dated phylogenies for bees (Trunz et al. Reference Trunz, Packer, Vieu, Arrigo and Praz2016). Ultimately, these efforts to increase taxonomic knowledge can inform conservation efforts (Soltis and Gitzendanner Reference Soltis and Gitzendanner1999; Hajibabaei et al. Reference Hajibabaei, Singer, Hebert and Hickey2007; Goldstein and DeSalle Reference Goldstein and DeSalle2011). Important caveats to relying on DNA methods are that reference material (i.e., identified specimens that have been sequenced) must be accurately identified (Collins and Cruickshank Reference Collins and Cruickshank2013), and that species do not show high levels of intraspecific variation (e.g., Spooner Reference Spooner2009).
The primary objective of this study is to summarise our efforts to date in developing a “Bees of Canada” DNA barcode database to facilitate taxonomic and faunal studies of bees in Canada. This effort builds on previous DNA-facilitated taxonomic revisions (e.g., Gibbs Reference Gibbs2010; Sheffield et al. Reference Sheffield, Ratti, Packer and Griswold2011b) and regional data sets (e.g., Sheffield et al. Reference Sheffield, Kevan, Smith, Rigby and Rogers2003, Reference Sheffield, Hebert, Kevan and Packer2009) for Canada, and other regional faunas or taxonomic treatments outside of North America (Carolan et al. Reference Carolan, Murray, Fitzpatrick, Crossley, Schmidt and Cederberg2012; Magnacca and Brown Reference Magnacca and Brown2012; Francoso and Arias Reference Francoso and Arias2013; Schmidt et al. Reference Schmidt, Schmid‐Egger, Morinière, Haszprunar and Hebert2015). Specifically, we will determine how well the accumulated number of unique barcode index numbers, which show high concordance with species (Ratnasingham and Hebert Reference Ratnasingham and Hebert2013) for sequenced Canadian bees, matches our numerical species tally and the known composition of species from Canada. Barcode index number assignment can be used to verify species identifications as well as document diversity when taxonomic information is lacking (Ratnasingham and Hebert Reference Ratnasingham and Hebert2013). Comparisons will be done nationally, provincially/territorially for species, and at the genus level. We also highlight how DNA barcoding is contributing to bee faunistic knowledge and traditional taxonomic work in Canada, and provide examples of how DNA barcodes can be, and have been, used to complement morphological methods to increase our taxonomic knowledge, including in phylogenetic studies, and discuss the issues and limitations with the current status of Canadian bee taxonomic knowledge. Thus, the diversity and taxonomy of bees in Canada provides a good illustration of the advances in biodiversity study that have occurred during the time period covered by the Biological Survey of Canada, and will complement the bees of Canada image library (www.yorku.ca/bugsrus/resources/galleries/boc), a checklist of the bees of Canada (in preparation) and a soon to be launched “Bees of Canada” website (www.beesofcanada.com). In addition to a full catalogue treatment, the latter will provide images, distributional information (i.e., jurisdiction, ecozone), floral hosts, and literature for all bee species occurring in Canada.
Methods
Material for DNA barcoding
The development of a DNA barcode library for bees began at a regional level (i.e., Nova Scotia) in the early 2000s, with a summary publication in 2009 (Sheffield et al. Reference Sheffield, Hebert, Kevan and Packer2009). Since the onset of the campaign to barcode Canada’s bees in 2006 – a component of a larger effort to barcode the world’s bees (Packer et al. Reference Packer, Gibbs, Sheffield and Kevan2008, Reference Packer, Sheffield, Gibbs, de Silva, Best and Ascher2009b) – samples have been collected from across North America, with special focus within southern Canada (Fig. 1). These have been combined with museum specimens from the following Canadian and United States of America collections: Packer Collection at York University; Biodiversity Institute of Ontario, University of Guelph (Guelph, Ontario, Canada); J.B. Wallis-R.E. Roughley Museum of Entomology, University of Manitoba (Winnipeg, Manitoba, Canada); Royal Saskatchewan Museum (Regina, Saskatchewan, Canada); Royal British Columbia Museum (Victoria, British Columbia, Canada); Simon Fraser University (Vancouver, British Columbia, Canada); Canadian National Collection of Insects, Arachnids, and Nematodes (Ottawa, Ontario, Canada); American Museum of Natural History (New York, New York, United States of America); Smithsonian National Museum of Natural History (Washington, District of Columbia, United States of America); United States Department of Agriculture, Agricultural Research Service Bee Biology and Systematics Laboratory (Logan, Utah, United States of America); United States Geological Survey Patuxent Wildlife Research Center (Laurel, Maryland, United States of America); and other contributors. The majority of DNA barcoded specimens from Canada are deposited at York University and the Biodiversity Institute of Ontario.
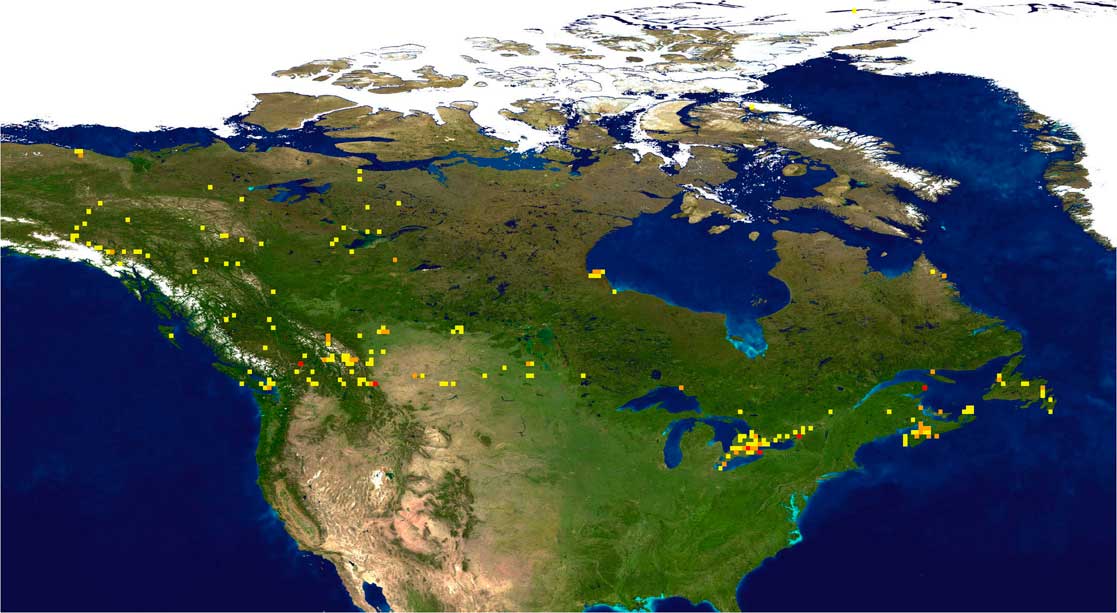
Fig. 1 Bee barcode sampling intensity in Canada. Darker coloured squares represent areas with more intensive sampling.
Tissue sampling and molecular protocols
For barcoding bees, tissue samples (i.e., usually a single mesothoracic or metathoracic leg from pinned specimens) were removed and sent to the Biodiversity Institute of Ontario for extraction and sequencing using well-established protocols (e.g., Hajibabaei et al. Reference Hajibabaei, Ivanova, Ratnasingham, Dooh, Kirk, Mackie and Hebert2005). Barcode index numbers are assigned to sequences in the Barcode of Life Datasystems (Ratnasingham and Hebert Reference Ratnasingham and Hebert2007) using a sequential process of algorithms using predefined distance thresholds with refined clustering of sequences, each of which represents an algorithmically grouped barcode sequence or group of sequences, with constituents of each barcode index number usually showing a high concordance within species boundaries (Ratnasingham and Hebert Reference Ratnasingham and Hebert2013). To summarise the DNA barcodes of the bee fauna of Canada as a whole, single representatives of each species/barcode index number, regardless of how many taxon names were associated with that barcode index number, were selected for analysis from the thousands of specimens that are currently barcoded. This differed from studies looking at intraspecific sequence divergence (which use multiple individuals of each species from multiple locations; Bergsten et al. Reference Bergsten, Bilton, Fujisawa, Elliott, Monaghan and Balke2012). Among this material, sequence data were downloaded from the barcode of life data system and imported in Mega Version 7 (Kumar et al. Reference Kumar, Stecher and Tamura2016) for sequence alignment and construction of an neighbour-joining tree using the neighbour-joining algorithm with the K2P model, with pairwise deletion of missing data, and the inclusion of all codon positions and substitution types (as used in the barcode of life data system analytical module). The patterns were then visualised using FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
A checklist of known Canadian bee species, including those in each province and territory was prepared for a national conservation assessment (Canadian Endangered Species Conservation Council 2016). This species checklist formed the basis for assessing completion of the DNA barcoding campaign for Canada. Genus-level summaries of the number of valid species known to occur in Canada, and comparable number of unique barcode index numbers were prepared. As the species data and barcode index numbers were also partitioned by jurisdiction (i.e., province and territory), tallies of barcode index numbers represented in each province (and associated with taxonomic information) were compared with these data to show diversity of both known species and barcode index numbers per jurisdiction. This included specimens within a single barcode index number, which had no species-level identification. Only one representative of each barcode index number was added to the tally, regardless of the species-level identification associated with the DNA barcoded specimens, as the comparisons were of known species to barcode index numbers within each genus.
Results and discussion
Much of the sampling to date has been focussed in areas of southern Canada (south of 50°N) (Fig. 1), but this region supports almost all of the species known from Canada (see Sheffield et al. Reference Sheffield, Frier and Dumesh2014), suggesting that the specimens received are largely representative of bee diversity in Canada. Approximately 14 200 bee specimens have been processed for DNA barcoding, and these have yielded 12 600 barcode-compliant sequences, resulting in 811 distinct barcode index numbers from within Canada (Fig. 2). This value represents 95% of the total of 856 species presently known from Canada (Table 1), but comparison of barcode index numbers with known species patterns in different genera shows that many more species may be present. It is likely that the actual number of species, and barcode index numbers assigned to these species, will continue to increase with continued sampling.
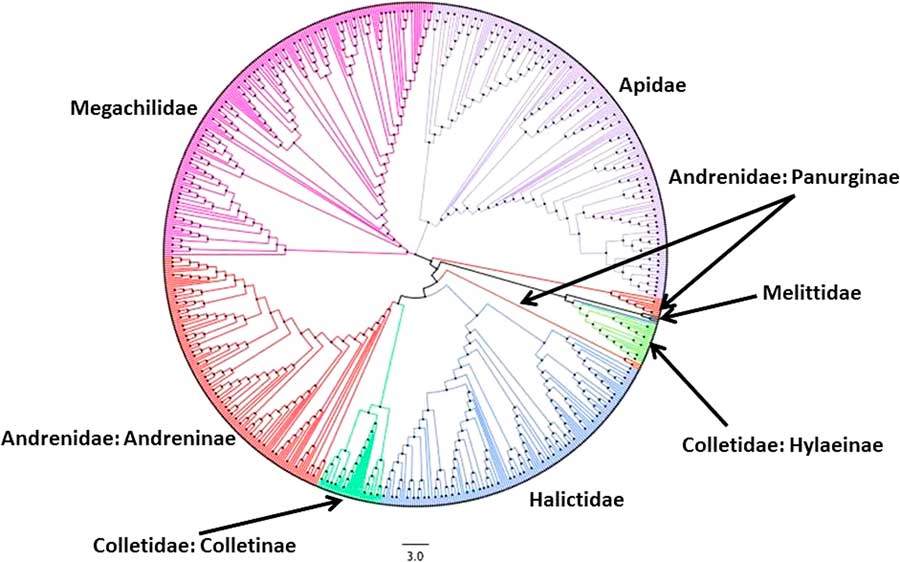
Fig. 2 Neighbour-joining (NJ) tree for single representative COI barcode sequences (BINs) for the bees of Canada. Colours represent the six bee families in Canada. Note that in this NJ tree, the families Andrenidae and Colletidae appear multiple times.
Table 1 Summary of the bee species in Canada with the number of species known and barcoded from each genus.

“ssp”=multiple subspecies are recognised in Canada. See generic treatment for genera with a “+”, “−”, or “*” in the last column; values in [] represent the difference when subspecies are considered.
Of the 52 genera currently recorded in Canada, only 25 (45%) have all known species barcoded, though most of these are represented by less than five species (Table 1, Figs. 3–4). In most (20) of the remaining genera, fewer barcode index numbers have been recorded than the known morphological species suggesting that these taxa have not been fully sampled. Some of these discrepancies are due to multiple morphologically distinct species sharing a barcode index number (e.g., Lasioglossum Curtis (Halictidae); see full discussion under Issue 2, below). By contrast, other genera have more barcode index numbers recorded for them than the known species in Canada (Table 1, Figs. 3, 4) even though two of these (Megachile Latreille (Megachilidae), Bombus Latreille (Apidae)) have had recent taxonomic treatment (Sheffield et al. Reference Sheffield, Ratti, Packer and Griswold2011b; Williams et al. Reference Williams, Thorp, Richardson and Colla2014, respectively). The remaining genera in this category are among those needing the most taxonomic work, and are largely responsible for the differences in the number of species and barcode index numbers observed in many parts of Canada (Fig. 5), especially British Columbia, the Prairie Provinces, and Ontario.
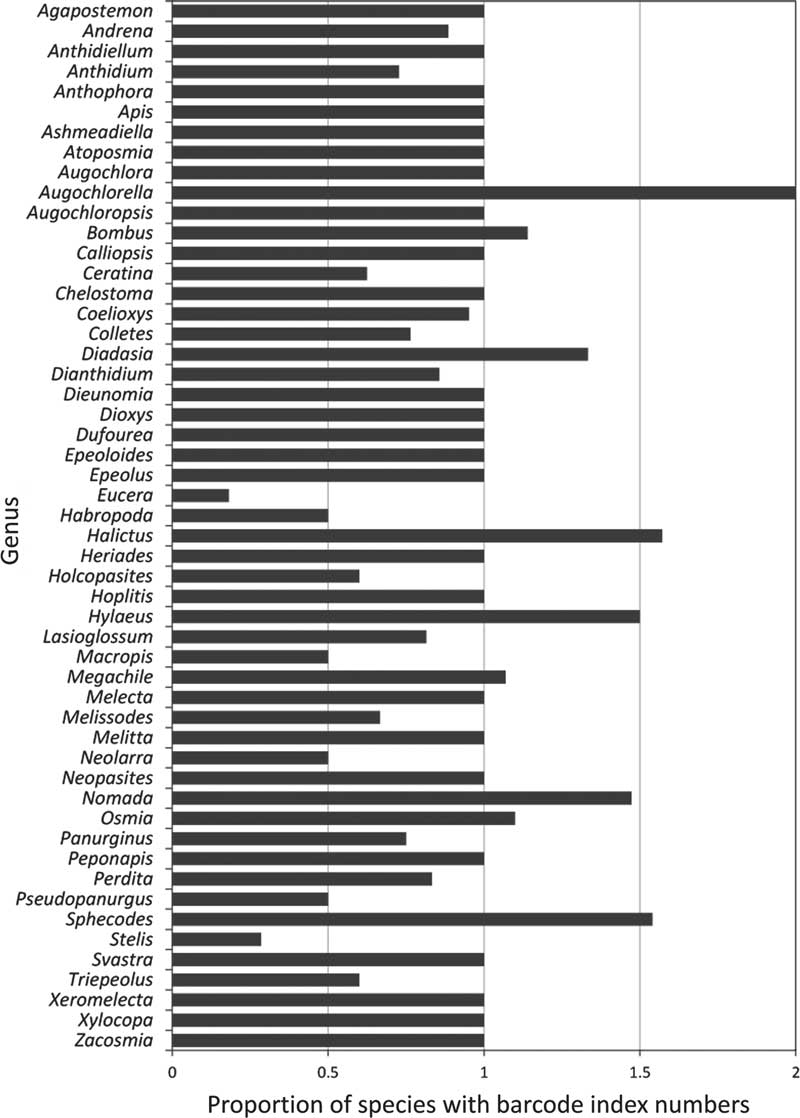
Fig. 3 Summary of the proportion of bee species for each genus for which there are barcodes with assigned barcode index numbers. Genera represented by bars less than 1.00 have more species recorded than barcode index numbers; greater than 1 have more barcode index numbers than species recorded in Canada.
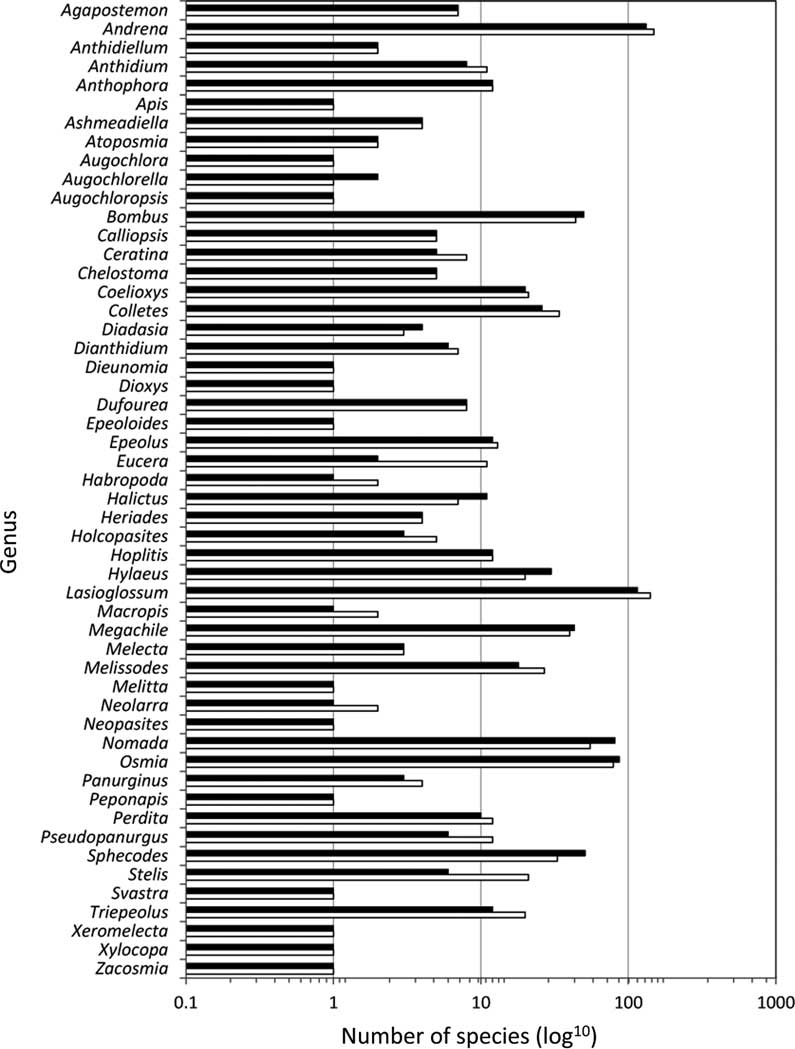
Fig. 4 The number of species (white bars) and number of barcode index numbers (black bars) for each genus of bee within Canada; x-axis log10 scale.
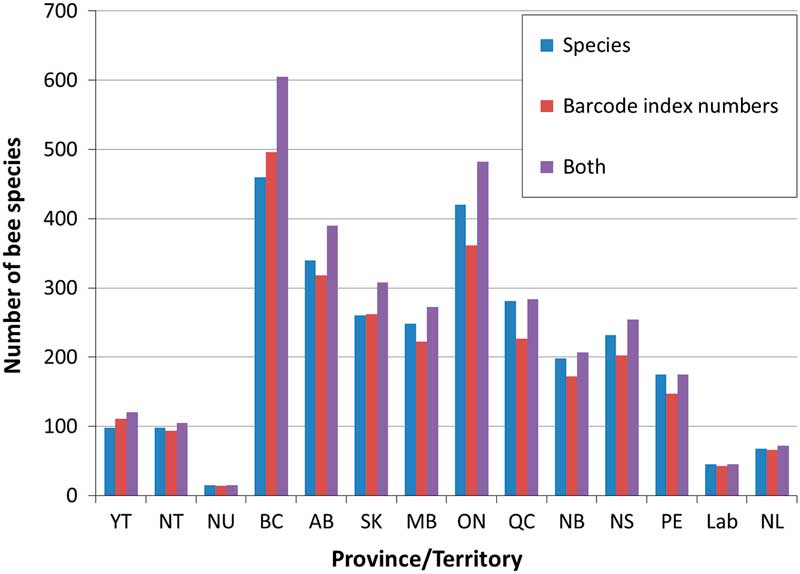
Fig. 5 The number of recorded bee species (blue bars) and barcode index numbers (red bars) for each jurisdiction in Canada. The combined data for both (purple bar) includes species and barcode index numbers, including barcode index numbers not associated with species names, so likely represents an overestimation of total species. YT, Yukon; NT, Northwest Territories; NU, Nunavut; BC, British Columbia; AB, Alberta; SK, Saskatchewan; MB, Manitoba; ON, Ontario; QC, Québec; NB, New Brunswick; NS, Nova Scotia; PE, Prince Edward Island; Lab, Labrador; NL, Newfoundland.
At the species level, of the 856 species recorded from Canada, 253 have no clearly associated barcode index number due to lack of material, poor success in obtaining sequences, or taxonomic uncertainty in assigning a name to the specimen associated with a sequence. An additional 28 species share barcode index numbers with other taxa, so the number of barcode index numbers cannot equal the number of species. On the other hand, 237 barcode index numbers are not currently associated to specific taxa (four of these represent cases where subspecific differences in barcode index numbers are observed).
The combined use of species and barcode index numbers alters our understanding of bee diversity in Canada, both nationally and at the provincial/territory level (Fig. 5). This is particularly true in British Columbia where the actual diversity of bees may fall between 500 and 600 species. These data suggest that there is still much taxonomic work to be done on Canadian bees, particularly within the genera Hylaeus Fabricius (Colletidae), Nomada Scopoli (Apidae), Sphecodes Latreille (Halictidae), and Osmia Panzer (Megachilidae), though a solid framework supporting taxonomic studies has been built in the barcode of life data system.
One issue to be resolved will be to ensure that all specimens sharing barcode index numbers are examined using traditional morphological taxonomic methods to support accepted taxonomy and nomenclature. Though specimens used to populate the bees of Canada DNA barcode project in barcode of life data system should be identified before being added (e.g., Collins and Cruickshank Reference Collins and Cruickshank2013), some speciose taxa are lacking keys and/or taxonomic specialists (and may have high numbers of undescribed species), making a priori identifications unlikely. Another consideration is the fact that many bee species are sexually dimorphic, and one of the sexes for many of these may be unknown or not associated (e.g., Sheffield and Westby Reference Sheffield and Westby2007), leading to inaccurate estimates of total species richness. Therefore, accessible DNA-based identification tools can be considered a major innovation in species diversity assessment in the 40 years since the inception of the Biological Survey of Canada. However, even as we should not assume that all taxa can be separated morphologically (e.g., Carman and Packer Reference Carman and Packer1996; Danforth et al. Reference Danforth, Mitchell and Packer1998; Packer et al. Reference Packer, Grixti, Roughley and Hanner2009a), neither can we assume that barcodes alone will address all biodiversity questions (Gibbs Reference Gibbs2010). This concept can be illustrated by examining the discrepancies between the known species and the barcode index numbers found in the bees of Canada data set. We summarise these below, provide a genus-by-genus account of where additional taxonomic work is required, and lastly provide examples of how and where combining DNA barcoding and traditional morphological taxonomy has been successful in studies of Canadian bees.
What are the issues?
Issue 1. One species: multiple barcode index numbers
Collins and Cruickshank (Reference Collins and Cruickshank2013) note that it is desirable to populate barcode of life data system with a priori identified specimens to build taxon-specific projects, though multiple barcode index numbers and/or high levels of sequence variation can be recognised after the fact. There are a few cases in which bee specimens recognised as good morphologically identified species have multiple barcode index numbers. For instance, among the well-studied bumble bees (Apidae: Apinae: Bombini), Bombus impatiens Cresson and B. ternarius Say each have three barcode index numbers represented by Canadian material in barcode of life data system. The fact that the representatives of each barcode index number cluster together suggests that this is not the result of misidentification but rather slight, but consistent variation in COI, without any pattern associated with morphology or geography.
Other discrepancies are likely due to cryptic species within poorly studied groups, or groups needing further study (see Discussions below). Another possibility is heteroplasmy, in which a single specimen can yield two different DNA barcodes (and possibly barcode index numbers), as reported for the genus Hylaeus by Magnacca and Brown (Reference Magnacca and Brown2009, Reference Magnacca and Brown2010), though this may largely go undetected as usually only a single tissue sample (e.g., a leg) is used for each specimen. Currently, studies have begun to address these types of patterns within the genus Hylaeus in Canada. At least three species, Hylaeus coloradensis (Cockerell), H. mesillae (Cockerell), and H. modestus Say show multiple barcode index numbers, though this group is rather difficult taxonomically, and much morphological variation within species has resulted in many synonymies. For example, H. modestus has two recognised subspecies and eight additional junior synonyms (Hurd Reference Hurd1979). These examples may also illustrate that for widespread species, it is important to sample multiple specimens of each species across its range; Bergsten et al. (Reference Bergsten, Bilton, Fujisawa, Elliott, Monaghan and Balke2012) suggest that at least 70 individuals from throughout the range (of a wide ranging species) would be required to account for 95% of the variation within a species. This may be even more important for Holarctic species; Andrena barbilabris (Kirby) (Andrenidae) is an example of a Holarctic species with three barcode index numbers in North America alone. Halictus confusus Smith (Halictidae) is an even more pronounced example, with five barcode index numbers in North America, though two subspecies are recognised on the continent, which may explain this partially, though Rosenmeier and Packer (Reference Rosenmeier and Packer1993) found no species-level differences among H. confusus populations from Alberta, Ontario, and Nova Scotia using electrophoretic methods for 40 loci. Sheffield and Perron (Reference Sheffield and Perron2014) offered some discussion of the implications for nomenclature for this species. There are other recent examples of Holarctic species being recognised as multiple taxa (e.g., Gibbs et al. Reference Gibbs, Packer, Dumesh and Danforth2013), including in the genus Bombus subgenus Alpinobombus Skorikov, in which multiple Holarctic species were recognised as separate Old and New World taxa (Williams et al. Reference Williams, Byvaltsev, Cederberg, Berezin, Ødegaard and Rasmussen2015); Williams et al. (Reference Williams, Cannings and Sheffield2016) later described one of these as a new species, B. kluanensis Williams and Cannings.
Issue 2. Multiple species: one barcode index number
In contrast to species with multiple barcode index numbers discussed above, there are also several cases in this data set in which multiple species share a single barcode index numbers. Examples are found in the genera Ceratina, Lasioglossum, and Bombus. Rehan and Sheffield (Reference Rehan and Sheffield2011) recently described a new species of Ceratina (subgenus Zadontomerus Ashmead) in eastern North America, one of four species sharing a single barcode index numbers. However, phylogenetic analysis of the COI data combined with morphological (Rehan and Sheffield Reference Rehan and Sheffield2011) and ecological data (Vickruck et al. Reference Vickruck, Rehan, Sheffield and Richards2011) supported the recognition of all four species as valid. Similarly, some clearly recognised species of bumble bees share barcode index numbers, though additional molecular analysis has facilitated the recognition of valid Nearctic/Palaearctic forms (Williams et al. Reference Williams, Byvaltsev, Cederberg, Berezin, Ødegaard and Rasmussen2015) and new species (Williams et al. Reference Williams, Cannings and Sheffield2016). In other cases (discussed below), incidences of multiple species sharing barcode index numbers have resulted in synonymies, largely supporting that many bumble bees are colour variable (Williams et al. Reference Williams, Thorp, Richardson and Colla2014). Lastly, there are many species of Lasioglossum subgenus Dialictus Robertson that share a barcode index number. In one example, at least 16 species share one barcode index number and would not be readily separated by DNA barcoding alone (i.e., use of barcode index numbers), though they can be separated by morphology and geography (Gibbs Reference Gibbs2010).
Issue 3. Multiple specimens, no barcode index numbers
Despite our attempts to obtain full DNA barcodes, a number of species have not yielded fully compliant sequences with barcode index numbers assigned. These include Andrena carlini Cockerell, Calliopsis chlorops Cockerell, Lasioglossum athabascense (Sandhouse), L. coeruleum (Robertson), L. colatum (Vachal), L. pallidellum Ellis, and L. reasbeckae Gibbs. Similar issues were found for some species in other bee-barcode campaigns, often associated with the presence and coamplification of the bacterial endosymbiont Wolbachia Hertig (Rickettsiaceae) (Magnacca and Brown Reference Magnacca and Brown2012; Schmidt et al. Reference Schmidt, Schmid‐Egger, Morinière, Haszprunar and Hebert2015).
Some Canadian species are very rare, some known only from type material collected decades previously (e.g., holotype and two paratypes of Andrena fulgida LaBerge), so it is likely that a DNA barcode library will never be complete unless these species are deemed synonymies of other taxa. It is important to note that there has been success in getting sequences from old material (Hajibabaei et al. Reference Hajibabaei, Smith, Janzen, Rodriguez, Whitfield and Hebert2006; Shokralla et al. Reference Shokralla, Zhou, Janzen, Hallwachs, Landry, Jacobus and Hajibabaei2011), though this is also influenced by methods used to collect and/or store specimens (Sheffield et al. Reference Sheffield, Hebert, Kevan and Packer2009). The willingness of institutions to have historic and rare type material subject to tissue removal for DNA barcoding is also likely an issue. Further, the rarity and/or taxonomic ambiguity of some of these species increases the probability that they will not be routinely sampled. As discussed by Lim et al. (Reference Lim, Balke and Meier2012), the sampling effort required to obtain some rare species is enormous.
Despite some of the difficulties mentioned above, many of which should be resolved with further molecular work and analyses (e.g., Dowton et al. Reference Dowton, Meiklejohn, Cameron and Wallman2014; Williams et al. Reference Williams, Byvaltsev, Cederberg, Berezin, Ødegaard and Rasmussen2015, Reference Williams, Cannings and Sheffield2016) and increased sampling (Bergsten et al. Reference Bergsten, Bilton, Fujisawa, Elliott, Monaghan and Balke2012), the value of DNA barcoding to support traditional taxonomic work has been illustrated in several works (see above and Collins and Cruickshank Reference Collins and Cruickshank2014). Analysis of the Canadian bee fauna provides a good example of this for some groups, and also illustrates where more taxonomic work needs to be done. We discuss the genera with discrepancies between the number of species and the number of barcode index numbers below (arranged by family and alphabetically by genus within each family) (Table 1).
Family Colletidae
Colletes Latreille. At present, 34 species are known from Canada, four of these with recognised subspecies. Specimens have not been collected and/or COI sequences have not yet been obtained for eight of these. However, Colletes kincaidii Cockerell is represented by two unique, yet closely associated barcode index numbers.
Hylaeus Fabricius. There are 30 unique barcode index numbers recorded from Canada, potentially representing ten more species than the 20 presently recorded from Canada. The taxonomy and identification of Hylaeus has traditionally been based on the presence and extent of colour markings, and variations within species have resulted in multiple junior synonyms for some (e.g., H. mesillae). In other species (e.g., H. affinis (Smith), H. modestus), females are very difficult to reliably distinguish. Members of this genus are also easily introduced outside of their native range, with several non-native species now established in North America, with many recent arrivals (Sheffield et al. Reference Sheffield, Dumesh and Cheryomina2011a; Gibbs and Dathe Reference Gibbs and Dathe2017; Martins et al. Reference Martins, Normandin and Ascher2017). Thus, undocumented non-native species are also a possibility. Magnacca and Brown (Reference Magnacca and Brown2009, Reference Magnacca and Brown2010) indicated that heteroplasmy within some Hylaeus (species outside of North America) creates some issues for successful specimen identification using DNA barcodes (i.e., potentially multiple barcode index numbers for a single specimen), though it is not presently known if this is an issue within the Canadian fauna as typically only a single tissue sample is used for each specimen.
Family Andrenidae
Andrena Fabricius. There are likely many more Andrena species yet to be recorded in Canada, in addition to the 149 species confirmed, 132 barcode index numbers have been recorded, 19 of which have not yet been examined for identification to species level. Several species have multiple barcode index numbers, including Andrena barbilabris (a Holarctic species represented by three barcode index numbers in North America alone). Andrena still requires much attention, as many misidentifications within barcode of life data system have resulted in multiple names associated with barcode index numbers.
Calliopsis Smith. Five species and five barcode index numbers have been recorded from Canada, though two of the barcode index numbers are from a single species, Calliopsis andreniformis Smith. Calliopsis chlorops has not yet been successfully barcoded, despite numerous attempts.
Panurginus Nylander. Four species are known from Canada, and only two have been barcoded with barcode index numbers. One species, P. ineptus Cockerell has two barcode index numbers. An additional specimen with a non-barcode-compliant sequence may represent a fifth species.
Perdita Smith. A total of 12 species have been recorded from Canada, with one represented by two subspecies. There are 10 barcode index numbers, two of these not yet associated with a named species.
Pseudopanurgus Cockerell. Of the 12 species recorded from Canada, two have associated barcode index numbers, with five additional barcode index numbers not yet assigned to a species.
Family Halictidae
Augochlorella Sandhouse. Coelho (Reference Coelho2004) synonymised A. striata (Provancher) under A. aurata (Smith) resulting in one species recorded from Canada, but there are two barcode index numbers. Ordway (Reference Ordway1966) originally suggested much morphological variation in A. striata, so it is likely that more detailed analysis of morphology and barcoding is required.
Halictus Latreille. At present, seven species are known from Canada, represented by 11 barcode index numbers. As discussed above, the Holarctic species H. confusus is responsible for much of the discrepancy (four barcode index numbers) (also see discussion in Sheffield and Perron (Reference Sheffield and Perron2014)), with H. tripartitus Cockerell accounting for two barcode index numbers.
Lasioglossum Curtis. The genus Lasioglossum has 141 known Canadian species and is represented by 115 barcode index numbers. Many of the species currently recognised morphologically (see Gibbs Reference Gibbs2010) share barcode index numbers and account for much of the discrepancy between barcode index numbers and species, with most of the remaining discrepancy due to the number of species not yet barcoded. There are cases, such as L. cressonii (Robertson) and L. ruidosense (Cockerell), where morphologically and geographically defined species have multiple barcode index numbers (Gibbs Reference Gibbs2010). The recent works of Gibbs (Reference Gibbs2009, Reference Gibbs2010, Reference Gibbs2011) and Gibbs et al. (Reference Gibbs, Packer, Dumesh and Danforth2013) are examples of the benefit of including DNA barcodes as part of revisionary taxonomy to resolve issues in difficult bee taxa.
Sphecodes Latreille. Sphecodes is one of the genera in Canada in most need of revision. At present, 33 species have been recorded from Canada, though there are 51 barcode index numbers. Currently, no key to species exists for western North America, and that for the east (Mitchell Reference Mitchell1960) is not representative of all the species in the area.
Family Melittidae
Macropis Panzer. Two species occur in Canada, both oligoleges of Lysimachia Linnaeus (Primulaceae) flowers, though one (M. ciliata Patton) is very rare, and no material has been barcoded.
Family Megachilidae
Anthidium Fabricius. At present, 11 species of Anthidium (three are non-native), are known from Canada, and eight of these have barcode index numbers.
Ashmeadiella Cockerell. There are four species recorded from Canada, one with two subspecies, and four barcode index numbers. It is likely that the DNA barcodes do not differ in the two subspecies.
Coelioxys Latreille. There are 21 species confirmed from Canada, and 20 barcode index numbers, though two species have no barcode index numbers associated with them, and two other species have multiple barcode index numbers. There may be misidentifications in barcode of life data system, but this group needs revision.
Dianthidium Cockerell. There are seven species recorded from Canada, and six barcode index numbers; two species (D. curvatum (Smith) from British Columbia, and D. simile (Cresson) from Ontario) share a barcode index number.
Hoplitis Klug. For the 12 species recorded in Canada, there are 12 barcode index numbers, but one species has not yet been barcoded. Though two species occurring in Canada have subspecies, no variation in barcodes is apparent within these species. Two taxa have multiple barcode index numbers.
Megachile Latreille. Since Sheffield et al. (Reference Sheffield, Ratti, Packer and Griswold2011b) revised the 38 species in Canada, Bzdyk (Reference Bzdyk2012) and Sheffield and Genaro (Reference Sheffield and Genaro2013) raised two subspecies to species (M. snowi Mitchell and M. cleomis Cockerell, respectively), resulting in 40 species in Canada. In all, 43 barcode index numbers have been recorded, but one species, M. umatillensis (Mitchell) has no barcode index number associated with it, M. gemula Cresson and M. relativa Cresson each have two, and M. pugnata Say has three (supporting subspecies).
Osmia Panzer. Due to the species richness of this genus, and the difficulty in identifying species, Osmia is one of the genera most in need of revision. Currently, there are 79 species recorded from Canada, and 87 barcode index numbers. Only 66 species have associated barcode index numbers, and there are 21 barcode index numbers not yet assigned to specific taxa and/or representing multiple barcode index numbers of a single named taxon. Likely many of these are the result of misidentification.
Stelis Panzer. These cleptoparasites are in need of revision, with 21 species recorded from Canada but only six with associated barcode index numbers.
Family Apidae
Anthophora Latreille. There are 12 species recorded from Canada, and 12 barcode index numbers, though three species are without barcode index numbers, and three have two barcode index numbers each. One from the latter group likely represents a misidentification.
Bombus Latreille. There are 44 bumble bee species recorded from Canada, including one new species described (Williams et al. Reference Williams, Cannings and Sheffield2016) since the last taxonomic treatment of the genus (Williams et al. Reference Williams, Thorp, Richardson and Colla2014). The 51 barcode index numbers from Canada include some species with multiple barcode index numbers (see B. impatiens and B. ternarius discussion above), species with recognised subspecies (B. bifarius Cresson and B. occidentalis Greene), and two others currently undergoing further taxonomic study.
Ceratina Latreille. Eight species of Ceratina are known from Canada, represented by five barcode index numbers. The shared barcode index number of four eastern species of Ceratina (Zadontomerus) is discussed above.
Diadasia Patton. The three species in Canada are represented by four barcode index numbers. One species, Diadasia australis (Cresson), is represented by two closely associated barcode index numbers, one on the eastern side of the Rockies, one on the west.
Epeolus Latreille. There are 12 unique barcode index numbers for the 12 species recorded from Canada, although a 13th species (yet to be barcoded) has been also been recorded in the country (Romankova Reference Romankova2004). The Canadian species of Epeolus were recently revised by Onuferko (Reference Onuferko2017).
Eucera Scopoli. Of the 11 species thought to be in Canada, only six have been barcoded and have yielded barcode index numbers. It is therefore likely that some species of Eucera are rare in Canada, but material in the barcode of life data system identified as five different species share a single barcode index number. This genus requires revision.
Habropoda Smith. Only one of the two species known from Canada, Habropoda cineraria (Smith), has been barcoded.
Holcopasites Ashmead. Five species are known from Canada, and three of these have barcode index numbers.
Melissodes Latreille. Melissodes is one of the taxa in need of attention; there are 27 species reported from Canada (one with two subspecies), and 18 barcode index numbers, though six of these are not yet associated with valid species.
Neolarra Ashmead. Only one of the two species known from Canada, Neolarra vigilans (Cockerell) has been barcoded.
Nomada Scopoli. Nomada is one of the most taxonomically difficult groups of bees in North America, with few recent keys, and many species known from one sex. This genus is perhaps most in need of taxonomic revision. There are 81 barcode index numbers in barcode of life data system from Canada, representing 55 known species. Only 15 species have been associated with barcode index numbers (and some of these are likely misidentified), and 66 barcode index numbers are without names.
Triepeolus Robertson. There are 20 species recorded from Canada, represented by 12 barcode index numbers, two of these not associated with a species name.
Future prospects for DNA barcoding and bee taxonomic studies
Associating sexes
Among the bees, many speciose genera have high proportions of species known from one sex; Sheffield and Westby (Reference Sheffield and Westby2007) indicated that ~37% of North American leafcutter bees were known from one sex. In a recent revision of the genus Megachile for Canada, Sheffield et al. (Reference Sheffield, Ratti, Packer and Griswold2011b) associated the sexes for all 38 species known at that time, resulting in several synonymies. For instance, Megachile alamosana Mitchell, previously known only from the male, was synonymised with Megachile anograe Cockerell, which was known only from the female (Sheffield et al. Reference Sheffield, Ratti, Packer and Griswold2011b) (Fig. 6). In addition, the male of M. sublaurita Mitchell was described for the first time (Sheffield et al. Reference Sheffield, Ratti, Packer and Griswold2011b). Similarly, in the genus Sphecodes, Sheffield et al. (Reference Sheffield, Hebert, Kevan and Packer2009) associated Sphecodes carolinus Mitchell to S. coronus Mitchell, known from the female and male, respectively. Gibbs (Reference Gibbs2010) also made 14 sex associations for Lasioglossum (Dialictus) in Canada, in a study that combined both morphological and molecular techniques.
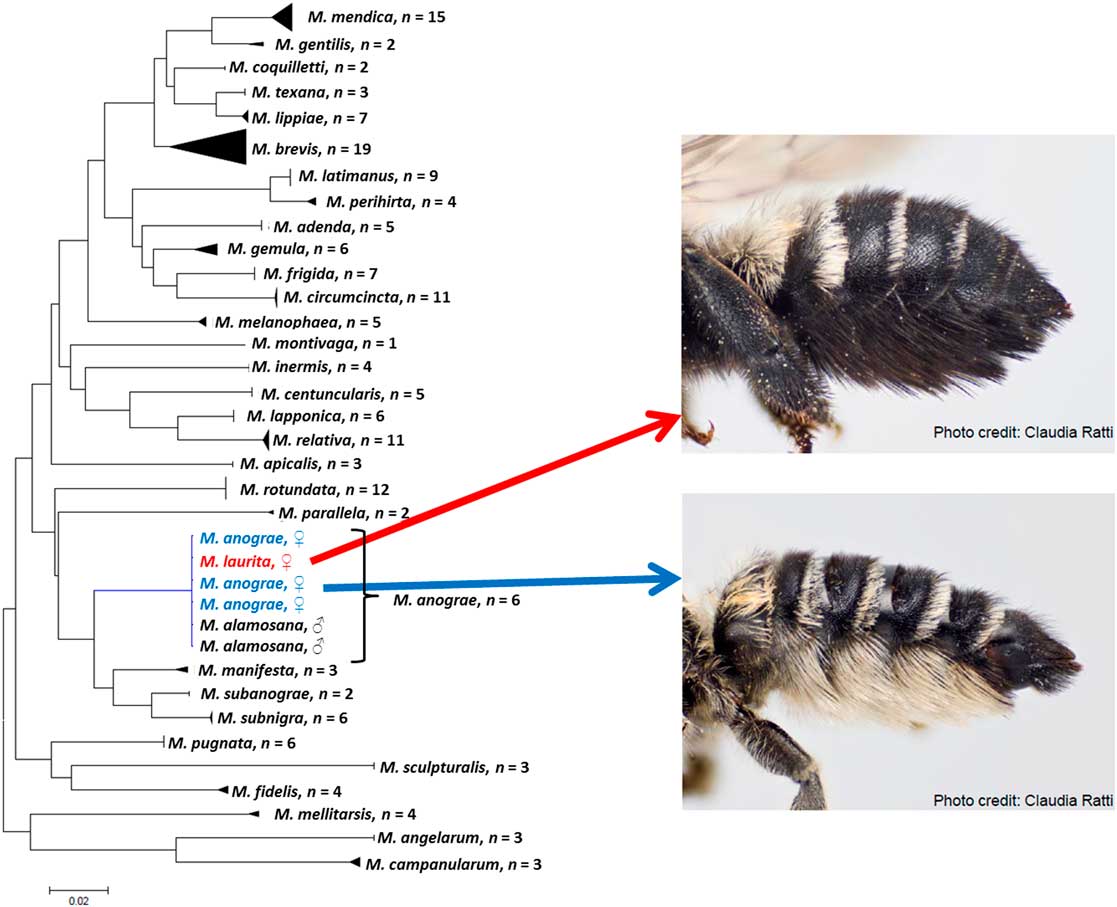
Fig. 6 A typical neighbour-joining tree for specimens of Megachile (Megachilidae) species in Canada. In this example, one clade is expanded to illustrate that DNA barcoding revealed that M. laurita and M. anograe (previously considered valid species known from the female only) were actually just colour variants of the same species. Also illustrated is the association of the male of M. anograe (M. alamosana, also previously considered a valid species). See Sheffield et al. (Reference Sheffield, Dumesh and Cheryomina2011a).
Morphological variation
In addition to being sexually dimorphic, some bee species also exhibit high morphological variability, especially colour. In bumble bees (genus Bombus) this is particularly common, though for some highly variable species (e.g., B. rufocinctus Cresson), no variation in COI has been reported, nor are there any obvious corresponding geographical patterns associated with these colour variants. In another example, Williams et al. (Reference Williams, Thorp, Richardson and Colla2014) considered B. californicus Smith a synonym of B. fervidus (Fabricius) due to little to no variation in COI (i.e., same barcode index number), though the darker form (i.e., B. californicus) is largely distributed in the west (including British Columbia and Alberta), though with intergrades occurring with the eastern yellow form (B. fervidus). However, despite the molecular evidence and synonymy of Williams et al. (Reference Williams, Thorp, Richardson and Colla2014), Dolan et al. (Reference Dolan, Delphia, O’Neill and Ivie2017) still recognised both species in Montana. In another series of studies with bumble bees, Williams et al. (Reference Williams, Brown, Carolan, An, Goulson and Aytekin2012) and Sheffield et al. (Reference Sheffield, Richardson, Cannings, Ngo, Heron and Williams2016) used geographic patterns of COI to recognise two subspecies of B. occidentalis, the latter study also including an analysis of geography-based colour variation to help define the distribution of each designatable unit, and assist in the Committee On the Status of Endangered Wildlife In Canada conservation assessment for each (Committee on the Status of Endangered Wildlife in Canada 2014).
Holarctic distribution and introduced species
Global DNA barcode initiatives, such as BeeBOL (Packer et al. Reference Packer, Gibbs, Sheffield and Kevan2008, Reference Packer, Sheffield, Gibbs, de Silva, Best and Ascher2009b) can help detect introduced species and clarify distributions. Introduced bees are commonly detected in Canada (Sheffield et al. Reference Sheffield, Richards and Griswold2010, Reference Sheffield, Dumesh and Cheryomina2011a; Gibbs and Dathe Reference Gibbs and Dathe2017; Martins et al. Reference Martins, Normandin and Ascher2017). More recently, DNA barcoding has helped clarify the distributional status of several bee species, including the confirmation of a Holarctic distribution in Bombus distinguendus Morawitz (Sheffield and Williams Reference Sheffield and Williams2011), and the recognition of separate Nearctic/Palaearctic distributions of bumble bees of the subgenus Alpinobombus (Williams et al. Reference Williams, Byvaltsev, Cederberg, Berezin, Ødegaard and Rasmussen2015).
Pollination studies
In addition to the benefits of incorporating DNA barcoding into traditional bee taxonomic studies, the opportunities for incorporating this technique into bee/pollinator ecological studies are many (Valentini et al. Reference Valentini, Pompanon and Taberlet2009; and see Vamosi et al. Reference Vamosi, Gong, Adamowicz and Packer2016 for a discussion related to pollinators and pollination). For instance, the development of techniques for DNA barcoding land plants (e.g., Kress et al. Reference Kress, Wurdack, Zimmer, Weigt and Janzen2005, Reference Kress, García-Robledo, Uriarte and Erickson2015; Kress and Erickson Reference Kress and Erickson2007; Hollingsworth et al. Reference Hollingsworth, Graham and Little2011; Li et al. Reference Li, Yang, Henry, Rossetto, Wang and Chen2015), and bee-collected pollen (e.g., Galimberti et al. Reference Galimberti, De Mattia, Bruni, Scaccabarozzi, Sandionigi and Barbuto2014) and/or honey (Valentini et al. Reference Valentini, Miquel and Taberlet2010) allow pollinating bees to be linked to their floral hosts in a range of ecosystems. However, pollen is not the only plant tissue used by bees – recently, MacIvor (Reference MacIvor2016) demonstrated how DNA barcoding could be used to identify the plant species that leafcutter bees (Megachildae) cut leaf pieces from to build their nests. Lastly, with the development of a DNA barcode library for bees, opportunities for associating cleptoparasitic bees to their host(s) using larvae excavated from nests would provide valuable information on bee communities (e.g., Sheffield et al. Reference Sheffield, Pindar, Packer and Kevan2013).
Acknowledgements
The authors thank Laurence Packer, York University, for his infectious obsession with bees, and for his support and encouragement for various studies of Canadian bees. Much of the initial financial support for graduate studies of Canadian bees came from Laurence through the Canadian Barcode of Life Network from Genome Canada (through the Ontario Genomics Institute), Natural Sciences and Engineering Research Council of Canada, and other sponsors listed at www.BOLNET.ca and via the National Science and Engineering Research Council (NSERC) Canadian Pollination Initiative (CANPOLIN). Additional funding for specimens in the south Okanagan provided by the British Columbia Parks, Park Enhancement Fund, British Columbia Ministry of Environment and federal Habitat Stewardship Program Prevention Stream to J.H.; the Saskatchewan Ministry of Agriculture and the Canada-Saskatchewan Growing Forward 2 bi-lateral agreement, and the Earth Rangers “Bring Back the Wild” campaign, both to C.S.S. Thanks are due to Paul D.N. Hebert, University of Guelph, for his early and continuing encouragement for bee DNA barcoding work, and the Biodiversity Institute of Ontario, University of Guelph for tissue sample preparation and all laboratory work associated with DNA barcoding. Lastly, the authors thank all the individuals and institutions that have provided specimens for use in their studies.









