INTRODUCTION
The genus Leptospira consists of a diverse group of pathogenic and saprophytic spirochaetes. Pathogenic members of this genus cause leptospirosis, a worldwide zoonotic disease, which affects man (Weil's disease, canicola fever, cane-field fever, 7-day fever, swineherd's disease) and animals (abortion, Stuttgart disease).
For epidemiological and epizootical purposes and also for diagnostic and therapeutic reasons, it is often necessary to detect the presence of pathogenic Leptospira in organs (particularly in renal tissue and urine, blood, cerebrospinal fluid) of infected humans and animals and determine its relevance with a particular leptospiral serovar/serogroup. This goal can be achieved by isolation of leptospiral strains and their identification by microagglutination tests (MATs) and cross-absorption tests [Reference Kmety and Dikken1]. In view of the fact that these classical methods are laborious and time-consuming and also require cultures of Leptospira strains as well as hyperimmune rabbit sera, their suitability is restricted to specialized laboratories.
The widely accepted basic systematic unit of leptospires has been the serovar defined on the basis of the structure of these microbes' antigens [2]. More than 200 serovars of pathogenic leptospires, categorized into 25 serogroups, have been described to date. Recently, eight genospecies of pathogenic leptospires have been differentiated on the basis of their DNA [2]. The two classification systems differ considerably – one genospecies often encompasses many serological very distinct serovars.
However, the serovar concept is important for epidemiological and public health reasons because different serovars may have different host animals and may to a certain extent be responsible for different clinical forms of the disease [2]. Therefore, numerous DNA-based methodologies have been elaborated in order to explore the genetic basis for the serovar affiliation of leptospiral strains [Reference Gerritsen, Smith and Olyhoek3].
Nowadays, the use of random amplified polymorphic DNA (RAPD) facilitates a rapid identification of isolated Leptospira strains because of its sensitivity, reliability, relative simplicity and speed [Reference Barocchi4–Reference Roy12].
However, the determination of the serogroup/serovar assignation of an unknown strain requires familiarity with the DNA banding patterns of Leptospira strains belonging to different serovars circulating in a given geographic region. This allows comparison of the DNA patterns of the new isolate with those of the representative serovars. Therefore, the RAPD method is thought to be suitable in cases when there is a limited number of serovars, which must be taken into account [Reference Brown and Levett5, Reference Corney, Colley and Graham6].
The aim of this study was to construct a phylogenetic tree of the leptospiral serovars encountered in Slovakia, Austria and Poland using calculations of DNA relatedness judged by the similarity of RAPD-generated DNA fingerprints. Furthermore, the usefulness of the phylogenetic tree (to compare antigenic properties with genetic relatedness) in comparing the DNA patterns of a new isolate with those of representative serovars and endemic strains isolated before 1991 was evaluated, in order to predict the serovar/serogroups' assignation of wild isolates of leptospires circulating not only in the aforementioned countries but, according to the literature [Reference Kathe and Mochmann13] and 50 years' experience of the WHO Collaborating Centre in Bratislava, in Central Europe as a whole.
MATERIAL AND METHODS
Reference strains of leptospires
The Leptospira strains listed in Table 1, with the exception of strain Vajany (endemic), are recognized by the Subcommittee on the Taxonomy of Leptospira as reference strains for different serovars [Reference Kmety and Dikken1]. They represent, according to our long-term observations and the relevant literature for Central Europe [Reference Kathe and Mochmann13], the most frequently found Leptospira serogroups/serovars in the above-mentioned regions. In this study, the endemic Leptospira strain Vajany will be also used as reference strain.
Table 1. Reference strains of serovars of leptospires representing Leptospira strains tested in this study

Leptospira strains recognized by the Subcommittee on the Taxonomy of Leptospira as reference strains of different serovars, with the exception of Vajany (endemic strain). Leptospira strains belonging to these serovars cause the majority of leptospirosis in Central Europe (according to the authors' experience and in the literature [Reference Kmety and Dikken1]).
Wild strains of leptospires
Ninety-one serotyped strains (n1) isolated between 1991 and 2002 and 50 non-serotyped strains (n2) of leptospires obtained during 2004 were subjected to genotyping. All these 141 Leptospira strains were isolated from different small terrestrial mammals, namely Apodemus flavicollis (yellow-necked field mouse), A. sylvaticus (long-tailed field mouse), A. agrarius (striped field mouse), A. microps (herb field mouse), Micromys minutus (harvest mouse), Mus musculus (house mouse), Clethrionomys glareolus (bank vole), Microtus arvalis (common vole), Pitymys subterraneus (pine vole) and Sorex araneus (common shrew). In order to verify that the genotyping method is of practical use, the serotyping of the latter mentioned 50 strains of leptospires followed their genotyping retrospectively.
Endemic strains isolated before 1991
For epidemiological purposes, the following endemic strains of leptospires of animal or human origin were also included in this study: OL-1 (serovar Mozdok), Myjava (serovar Grippotyphosa), Pöŝtényi (serovar Pomona), Biela myŝ (serovar Arborea), M-37 and Šaca (serogroup Sejroe), Petrík and PB-4 (serovar Icterohaemorrhagiae), Lebe, PB-3 and OL-2 (serovar Copenhageni) were considered and tested.
DNA preparation
All Leptospira strains used in this study were cultured in liquid Korthof and Ellinghausen–McCullough–Johnson–Harris (EMJH) media (Difco Laboratories, Detroit, MI, USA) and the bacteria harvested as described previously [Reference Awad-Masalmeh14]. Using the DNeasy® Tissue kit (Qiagen, GmbH, Hilden, Germany), DNA of each strain was extracted according to the manufacturer's instructions and the total amount of DNA of each strain was measured (Dyna Quant 200 Fluorometer; Hoefer Inc., San Francisco, CA, USA). To generate a valuable DNA fingerprint (7–8 bands) using the RAPD procedure, the concentration of the appropriate DNA target of each of the reference leptospiral strains was estimated using different DNA dilutions and, in comparison to this, the amount of target DNA from each wild strain to be tested was calculated.
RAPD testing was performed as described previously [Reference Awad-Masalmeh14] using the primer O5 5′-AGGGGTCTTG-3′ synthesized by VBC-Biotech Service GmbH (Vienna, Austria), and Ready-To-Go™ analysis beads [Amersham Biosciences, Bucks, UK (now part of GE Healthcare)]. Out of 20 primers, the O5 primer was selected by pretesting. The total volume (25 μl) of each reaction contained one Ready-To-Go™ bead, 2 μl target DNA solution (1·5–2 ng/μl), 2 μl primer O5 (concn 20 mm) and 21 μl sterile double-distilled water.
All amplification reactions were performed in a PerkinElmer Cycler 9600 (Norwalk, CT, USA) under the amplification conditions recommended by Williams et al. [Reference Williams15]: 94°C for 1 min, 40 cycles (94°C for 1 min, 36°C for 1 min, 72°C for 2 min), and 72°C for 3 min. PCR products were resolved by electrophoresis (100 V, 75 min; Bio-Rad castgel) using 2·5% agarose (Ultra pure probe agarose; Bio-Rad Laboratories, Inc., Hercules, CA, USA), stained with ethidium bromide and photographed; the digitally captured profiles were analysed using the Gel Doc™ 2000 system using Quantity One® Software (Bio-Rad). To achieve reproducible DNA fingerprints, the GeneRuler™ 100 bp DNA ladder Plus, no. SM0323 (Fermentas UAB, Vilnius, Lithuania) was applied to the outer lanes and, normally, also to three intermediate lanes of each agarose gel (Fig. 1).

Fig. 1. RAPD fingerprints of the strains representing the serovars of leptospires used. Lane M, ladder 100 bp+ MBI; lane 1, serovar Pomona strain Pomona; lane 2, serovar Lora strain Lora; lane 3, serovar Icterohaemorrhagiae strain RGA; lane 4, serovar not tested strain Vajany; lane 5, serovar Mozdok strain 5621; lane 6, serovar Djatzi strain HS 26; lane 7, serovar Grippotyphosa strain Moskva V; lane 8, serovar Sejroe strain M 84; lane 9, serovar Istrica strain Bratislava; lane 10, serovar Polonica strain 493 Poland; lane 11, serovar Saxkoebing strain Mus 24; lane 12, serovar Sorexjalna strain Sorex Jalná; lane 13, serovar Arborea strain Arborea.
Phylogenetic analysis of the DNA fingerprints was performed with the Bio-Rad Fingerprinting™ II software package (Fig. 2). Phylogenetic trees were constructed by complete linkage using the UPGMA clustering method with band matching by the Dice coefficient (Figs 2–4).

Fig. 2. DNA fingerprints and DNA relatedness to showing the connection between some representative wild isolates (†) and the corresponding reference strains (*) of Leptospira.

Fig. 3. DNA relatedness among Leptospira reference strains tested in this study. * Leptospira strains recognized by the Subcommittee on the Taxonomy of Leptospira as reference strains for different serovars with the exception of Vajaby (endemic strain). Leptospira strains belonging to these serovars cause, to the best our knowledge, the majority of leptospirosis in Central Europe (see also ref. [Reference Kmety and Dikken1]).

Fig. 4. DNA relatedness among Leptospira reference strains; wild strains (n1/n2) and endemic strains selected. * Reference strain (recognized by the Subcommittee on the Taxonomy of Leptospira). † Endemic strains isolated before 1991. ‡ (n1/n2), n1=serovar known before testing DNA relatedness; n2=serovar known after testing DNA relatedness.
Experimental strategy
The first step was to obtain DNA fingerprints and, from these, to create a dendrogram for the 13 reference strains of Leptospira (including the strain Vajany) representing the most frequently found serovars in our region (Table 1, Fig. 3). In the same manner, Figure 4 was also constructed using the DNA profiles obtained from the 141 wild isolates, of the 11 endemic and 13 reference strains of leptospires.
RESULTS
Each of the reference leptospiral strains used revealed a diverse, yet consistent and reproducible, DNA banding pattern (Fig. 1), and the phylogenetic tree, generated from these data, allowed us to distinguish between the 13 representative strains of eight serogroups of leptospires encountered in Central Europe. As indicated in Figure 3, all Leptospira serovars represented by their reference strains used remained distinguishable, despite a close DNA relationship found between Pomona and Lora, Mozdok and Vajany, Sejroe and Istrica, as well as between Saxkoebing and Sorexjalna.
Furthermore, the association between the achieved DNA fingerprints obtained from some representative wild isolates and from the corresponding serovar of the reference strains tested is presented in Figure 2.
The behaviour of the 141 wild isolates of leptospires compared to those of the reference strains tested, as well as to selected endemic ones, is presented in Figure 4. As shown, the tested wild strains of Leptospira serovars (34 Grippotyphosa, 34 Mozdok, 3 Pomona, 1 Sorexjalna) occupied the same position in the phylogenetic tree as the reference leptospiral strains of the particular serovars (Table 2a, Fig. 4). The 41 wild strains of serogroup Australis showed the same position as Lora, while the 27 strains of the serogroup Sejroe have been grouped into four clusters (12 with serovar Polonica, 11 with serovar Istrica, 3 with serovar Saxkoebing, and 1 with serovar Sejroe). In Figure 4, the strain RGA serovar Icterohaemorrhgiae and the strain M 20 serovar Copenhageni (both serogroup Icterohaemorrhagiae) occupied the same position, and similarly the members of serogroup Australis (strain Lora serovar Lora, strain Jež Bratislava serovar Bratislava, and strain Jalná serovar Jalná) remained indistinguishable.
Table 2 (a). Agreement at the serovar level with genotype (position in phylogenetic tree) of Leptospira wild strains
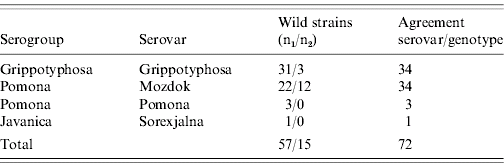
n1, Serovar known before testing DNA relatedness.
n2, Serovar known after testing DNA relatedness.
In contrast to this, Figure 4 illustrates the power to differentiate between the two members of serogroup Pomona (strain Pomona serovar Pomona and strain 5621 serovar Mozdok) and between those of serogroup Sejroe (strain M 84 serovar Sejroe, strain Bratislava serovar Istrica, strain 493 Poland serovar Polonica, strain Mus 24 serovar Saxkoebing).
Further, as indicated in Figure 4 and Table 2 b, the position of the wild isolate of serogroup Bataviae was found to be closer to Vajany than to that of the reference strain HS 26 serovar Djatzi of this serogroup.
Table 2 (b). Agreement at the serogroup level with genotype (position in phylogenetic tree) of Leptospira wild strains

n1, Serovar known before testing DNA relatedness.
n2, Serovar known after testing DNA relatedness.
Regarding the endemic Leptospira strains tested: OL-1 corresponds with serovar Mozdok; Myjava with Grippotyphosa; Pöštényi with Pomona; Biela myš with Arborea; Šaca with 493 Poland; M-37 with Sejroe; Petrík and PB-4 with Icterohaemorrhagiae; and Lebe, PB-3 and OL-2 with serovar Copenhageni (Fig. 4).
DISCUSSION
In this study, a phylogenetic tree of reference Leptospira strains representing the leptospires most frequently encountered in Central Europe [Reference Kathe and Mochmann13] has been constructed for the first time (Fig. 3); this tree was also found to be useful in distinguishing and identifying both wild and endemic strains of these bacteria. The correspondence between the unique position within the phylogenetic tree (genotype) and the serotype of each serovar of the 13 reference strains considered is shown based on to the results of repeated testing (Fig. 3).
These observations were also confirmed by the results of wild isolates of leptospires and by those of endemic strains with the exception of serovars belonging to serogroups Australis and Icterohaemorrhagiae, which were subjected to the same testing procedure. It was shown that the results of genotyping and serotyping at serovar/serogroup level of each of the 141 wild leptospiral strains of this study were in agreement [Fig. 4, Table 2(a and b)].
Furthermore, agreement between the results of genotyping and those of serotyping at the serovar level of wild strains (Table 2a) was achieved in 72 examined strains (belonging to serovars Grippotyphosa, Mozdok, Pomona, Sorexjalna).
It is noteworthy that the strains belonging to the Pomona and Mozdok serovars (both serogroup Pomona) can be serologically reliably distinguished only by using monoclonal antibodies, and not with polyclonal rabbit antisera used in classical MATs and cross-absorption tests.
At the serogroup level, the correspondence with genotyping was observed with 69 (41 Australis, 27 Sejroe, 1 Bataviae) leptospiral wild strains tested (Table 2b). The wild strains of Australis serogroup belonging to serovar Jalna (Fig. 4) remained indistinguishable from serovars Bratislava and Lora from the same serogroup. On the other hand, the 27 strains assigned to serogroup Sejroe form four different clusters according to the reference strains used in the case of this serogroup. Their identification at the serovar level is currently being performed.
The wild strain belonging to serogroup Bataviae presented more genetic relationship to the endemic strain Vajany than to the reference strain Djatzi. The serovar affiliation of strain Vajany has not been established yet. However, the position in Figure 4 of the wild isolate confirms the assignation of this strain to serogroup Bataviae.
At least in cases of the most frequently found Leptospira strains in our region, the presented results demonstrate the usefulness of the phylogenetic tree in predicting the serovar/serogroup of an unknown Leptospira strain from the genotype and vice versa, as well in distinguishing between endemic strains isolated before 1991.
These findings enhance and shorten the time of the identification of wild isolates of Leptospira, since the typing of these bacteria into serovars and/or serogroups by serological methods (i.e. by MATs and cross-absorption tests) is tedious and requires the maintenance of a comprehensive collection of strains as well as the corresponding rabbit immune sera [Reference Kmety and Dikken1, Reference Romero and Yasuda11].
The phylogenetic tree constructed during this study is based on the comparison, using different computer programs, of digitally captured and analysed RAPD-banding patterns of each leptospiral strain tested with each of those of the representative serovars; each of these banding patterns was diverse between strains, yet consistent and reproducible for each strain. This minimizes significantly the variations of visual reading of the DNA fingerprints, which have been used in previous studies [Reference Brown and Levett5–Reference Collares-Pereira7, Reference Ramadass9–Reference Roy12, Reference Tyler16–Reference Vijayachari18]. Furthermore, and in contrast to the studies cited above, the efficiency and accuracy of RAPD analysis in our study was increased by the use of manufactured amplification products [Ready-To-Go™ RAPD analysis beads; Amersham Biosciences (now part of GE Healthcare)], optimized primer and DNA template, the same amplification protocol and repeated testing in order to avoid day-to-day variance.
We therefore conclude that the results of this study indicate that the use of RAPD-based typing in conjunction with phylogenetic analysis are, at least for the Leptospira strains most frequently occurring in Central Europe (according to the WHO Collaborating Centre, Bratislava and in the literature [Reference Kathe and Mochmann13]), a potentially useful new tool in following occurrence, especially of outbreaks of leptospirosis by aiding the rapid identification of strains, a process necessary for containment strategies. One disadvantage of the method was the inability to distinguish between members of serogroup Australis, namely Jalna, Bratislava and Lora as well as the inability to distinguish between serovars Icterohaemorrhagiae and Copenhageni in serogroup Icterohaemorrhagiae. It is anticipated that this problem may be resolved using other primers. Nevertheless the results of this study represent the first step in establishing further investigations to gain more knowledge on virulence, host specificity, distribution and transmission of leptospires.
ACKNOWLEDGEMENTS
We thank Professor Baumgartner, head of the Clinic for Ruminants, the members of the Institute for Milk Hygiene, Milk Technology and Food Science, University for Veterinary Medicine, Vienna, and the technicians of the Institute of Epidemiology, Comenius University, Faculty of Medicine, Bratislava. Our special thanks go to Professor Iain Wilson for language advice. This work was financially supported by the Austrian Buiatric Society of the Austrian Veterinary Organisation and the Slovak Grant Agency VEGA, project no. 1/8293/01 and 1/1166/04.
DECLARATION OF INTEREST
None.









