Atlantic salmon (Salmo salar) is one of the most economically important aquaculture species constituting 35 % of global marine fish production( 1 ). The production of farmed salmon increased rapidly from 500 metric tons in 1970 to more than 2·3 million metric tons in 2014( 1 ). However, shortages of sufficient fishmeal and oils have led to massive substitutions (60 %) by plant ingredients in salmon diets( Reference Ytrestoyl, Aas and Asgard 2 ). This includes an increase in plant oils (PO) from 0 % in the diet in 1990 to 19·2 % in 2013( Reference Ytrestoyl, Aas and Asgard 2 ). As the lipid composition of PO is significantly different from that in fish oil (FO), this can influence growth and performance of salmon at early developmental stages.
The gastrointestinal tract of salmon consists of three major regions, the stomach, pyloric caeca and midgut, and hindgut, each of them with separate functions in lipid metabolism. Most studies on dietary lipid utilisation have focused on the pyloric caeca or midgut as they are the predominant regions for lipid absorption and transport in salmon( Reference Carmona-Antonanzas, Taylor and Martinez-Rubio 3 – Reference Tocher, Bell and McGhee 6 ). The hindgut could also be involved in absorption and transport of lipids, as both histological and genetical studies have suggested an existence of lipoproteins in the region( Reference Calzada, Medina and Canales 7 , Reference Jin, Olsen and Østensen 8 ). The stomach is primarily responsible for mechanical and chemical digestion of feed, but may also be involved in absorption of SCFA, Na and water( Reference Saunders 9 ). Similar to other teleosts, the stomach and pyloric caeca of salmon are the last organs to be completed during ontogenetic development of the digestive system( Reference Lazo, Darias and Gisbert 10 , Reference Sahlmann, Gu and Kortner 11 ). The stomach and hindgut are distinguishable 7 d after hatching, whereas the pyloric caeca occurs 20 d later( Reference Sahlmann, Gu and Kortner 11 ). Although the whole intestine of salmon is prepared for digestion and absorption of external feed before the yolk sac is depleted( Reference Sahlmann, Gu and Kortner 11 ), it is still unclear whether the capacities of digestion and absorption in salmon fingerlings are developed enough to tolerate a plant-based diet with high PO contents. The adult salmon can tolerate partial replacement of PO in diet without influencing growth( Reference Ytrestoyl, Aas and Asgard 2 , Reference Bligh and Dyer 12 – Reference Tocher, Fonseca-Madrigal and Bell 16 ). However, feeding PO could still cause massive accumulation of lipid droplets in enterocytes of the pyloric caeca, indicating abnormal lipid transport across the intestinal region( Reference Olsen, Myklebust and Kaino 4 , Reference Olsen, Myklebust and Ringo 5 , Reference Olsen, Dragnes and Myklebust 17 ). The liver is also important for synthesising and transporting lipids in salmon. Previous studies mostly used liver tissue to evaluate PO-induced changes of lipid metabolism in large salmon( Reference Tocher, Fonseca-Madrigal and Bell 16 , Reference Leaver, Villeneuve and Obach 18 ).
Like all vertebrates, salmon require cholesterol for growth and development as it is involved in many biological functions, such as formation of cell membranes and provision of bile acids( Reference Van der Wulp, Verkade and Groen 19 ). Cholesterol homoeostasis in the fish body is achieved by balancing dietary intake, de novo synthesis in the body and excretion as bile acids. PO is devoid of cholesterol, but contains several types of phytosterols( Reference Liland, Espe and Rosenlund 20 , Reference Szterk, Roszko and Sosińska 21 ). Although these phytosterols have lower bioavailability, they can further reduce the absorption of available dietary cholesterol( Reference Liland, Espe and Rosenlund 20 ). This would increase the demand on de novo synthesis, as shown by the up-regulation of cholesterol biosynthesis pathways in liver( Reference Leaver, Villeneuve and Obach 18 , Reference Gillard, Harvey and Gjuvsland 22 ). Whereas adult salmon are self-sufficient in cholesterol when on a PO diet( Reference Sissener, Liland and Holen 23 ), this ability in salmon fingerlings is largely unknown.
Atlantic salmon also require dietary supplement of long-chain PUFA (LC-PUFA, ≥C20) such as DHA (22 : 6n-3), EPA (20 : 5n-3) and arachidonic acid (ARA, 20 : 4n-6) for optimum growth and development( Reference Monroig, Navarro and Tocher 24 ). The requirement of LC-PUFA for salmon as for other fish is generally higher for fingerlings than juvenile or adult stages( Reference Tocher 25 – Reference Bou, Berge and Baeverfjord 27 ). However, as PO does not contain LC-PUFA, their content will be reduced upon feeding PO diets. Salmonids are capable of elongating and desaturating 18 : 3n-3 and 18 : 2n-6 to LC-PUFA( Reference Tocher 28 ), and fatty acyl desaturase (fads) and elongase (elovl) genes are often up-regulated in salmon upon PO feeding( Reference Tocher 15 ). However, the impact of elongation and desaturation on LC-PUFA level in salmon is often much less than that of diet( Reference Sargent, Bell and McEvoy 29 ). Furthermore, the capacity to elongate and desaturate is largely unknown in salmon fingerlings and early juveniles compared with larger individuals.
All salmonids have experienced an extra round of whole-genome duplication compared with other fish species. This salmonid-specific duplication (Ss4R) occurred in a common ancestor of all salmonids approximately 100–80 million years ago( Reference Macqueen and Johnston 30 , Reference Wright, Johnson and Hollister 31 ). To date, approximately 55 % of the Ss4R gene duplicates are still retained as expressed genes in Atlantic salmon( Reference Lien, Koop and Sandve 32 ). Some of the Ss4R duplicates have been found to have distinct regulation in different tissues( Reference Jin, Olsen and Østensen 8 ), developmental stages( Reference Jin, Olsen and Østensen 8 ) and dietary treatment( Reference Gillard, Harvey and Gjuvsland 22 ). This has markedly increased the complexity of understanding lipid metabolism pathways in salmon, meaning the functional divergence of gene duplicates must be taken into consideration in diet-induced gene expression studies.
In the present study, we applied transcriptomic and lipid analysis on the stomach, pyloric caeca, hindgut and liver of salmon fed either PO- or FO-enriched diets. The developmental aspect included first feeding (0·16 g), fingerling (2·5 g) and juvenile (10 g) salmon. The overarching goal was to achieve a systemic overview of PO-induced lipid metabolism changes in salmon fingerlings and juveniles. Expressional differences between Ss4R duplicate genes were also taken into consideration for studying the salmonid-specific features of genetic regulation of lipid metabolism.
Methods
Fish, diets and experimental plan
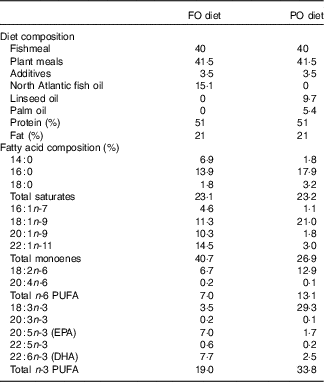
This study was carried out within the Norwegian animal welfare act guidelines, in accordance with EU regulation (EC Directive 86/609/EEC). Atlantic salmon eggs were hatched and cultivated at AquaGen Breeding Centre. The fish were fed either PO or FO diets (Cargill Innovation Centre) from first feeding up to 100 d. The PO used in this study was a mixture of palm and linseed oil. The composition of the two diets and their fatty acids are summarised in Table 1. A previous experiment used the same diets for studying the effect of PO on gene regulation in larger salmon before and after smoltification( Reference Gillard, Harvey and Gjuvsland 22 ). We sampled three fish from each dietary group at days 0 (0·16 g, before first feeding), 65 (2·5 g, fingerlings) and 100 (10 g, juvenile) after first feeding. The fish were euthanised by 1 g/l MS-222 (Finquel, Argent Chemical Laboratories, Inc.) buffered with the same amount of sodium bicarbonate before dissection in a sterile Petri dish filled with RNAlater. Samples of stomach, pyloric caeca, hindgut and liver were dissected, cleaned of connective tissues and intestinal contents and immediately transferred to 1 ml of RNAlater for RNAseq analysis. Tissues were stored for 24 h at 4°C for sufficient penetration of RNAlater before being transferred to −80°C for storage. Tissues of stomach, pyloric caeca and hindgut for lipid analysis were flash-frozen in liquid nitrogen and stored at −80°C.
Table 1 Composition of plant oil (PO) and fish oil (FO) diets
Lipid class and fatty acid analyses
Total lipids were extracted from salmon tissues using the method of Folch et al. ( Reference Folch, Lees and Sloane 33 ). Lipid classes were separated using the double development high-performance TLC method of Olsen & Henderson( Reference Olsen and Henderson 34 ). Extracted total lipid was applied onto 10×10 cm silica plates (Merck) for lipid class separation. The plates were developed using methyl acetate–isopropanol–chloroform–methanol–0·25 % KCl (25:25:25:10:9, by vol.) to separate polar lipids and hexane–diethyl ether–glacial acetic acid (80:20:2, by vol) for separation of neutral lipids. The phosphatidylcholine (PtdCho), phosphatidylethanolamine (PtdEtn) and TAG fractions were detected under UV light, scraped off and directly methylated with sulphuric acid–methanol (2:98, v/v) in isooctane at 50°C overnight. Isooctane and 5 % NaCl were then added to the mixture and centrifuged at 1640 g for 3 min at 4°C. The fatty acid methyl esters in the upper phase were analysed on an Agilent 7890B gas chromatograph with flame ionisation detector (Agilent Technologies). Fatty acid data were generated from two individual samples with two technical replicates (2×2).
RNA extraction and transcriptomic analysis
Three individual samples per tissue and experimental condition were used as biological replicates. Total RNA was extracted using RNeasy Plus Universal Kits (Qiagen). RNA concentration and purity were assessed by Nanodrop 8000 (Thermo Scientific) and RNA integrity was checked by Agilent 2100 Bioanalyzer (Agilent Technologies). Libraries were prepared by using TruSeq Stranded mRNA Library Prep Kit (Illumina), according to the manufacturer’s instructions. Samples were sequenced using 100-bp single-end high-throughput mRNA sequencing (RNA-Seq) on Illumina Hiseq 2500 (Illumina) in Norwegian Sequencing Centre. The raw sequences are publicly available on European Nucleotide Archive under accession number PRJEB21981. The raw sequence data for FO samples were used in a previous study to understand the transcriptional development of phospholipid and lipoprotein metabolism in salmon fry( Reference Jin, Olsen and Østensen 8 ).
Statistical analysis and data mining
Fatty acid analysis data were checked for statistical significance by using one-way ANOVA with Tukey’s multiple comparison test, and differences were considered significant when P<0·05.
For transcriptomic analysis, the quality control and counting of the read sequences were performed by the same method as described previously( Reference Gillard, Harvey and Gjuvsland 22 ). The uniquely mapped reads, aligned to exon regions, were counted for each gene in the salmon genome annotation (ICSASG_v2). Genes were filtered by a minimum count level of at least 1 count per million (CPM) in two or more samples, to remove genes with too few counts. For each tissue type (stomach, pyloric caeca, hindgut and liver), at each developmental stage (2·5 g fingerlings and 10 g juvenile), a differential expression analysis (DEA) was performed comparing PO with FO samples. For visually comparing expression levels between different genes and tissues, normalised counts in the form of transcripts per million values were generated. Raw gene counts were first divided by their mRNA length in kb to normalise for transcript length, and then divided by the total number of counts from each library to normalise for sequencing depth. For comparison of expression between Ss4R duplicated genes, an Ss4R duplicate gene list defined in a previous study was applied( Reference Gillard, Harvey and Gjuvsland 22 ). The present study had a special focus on genes involved in lipid metabolism pathways; therefore, 353 salmon lipid genes were selected from previous orthologue annotations to Kyoto Encyclopedia of Genes and Genomes (KEGG) database terms( Reference Gillard, Harvey and Gjuvsland 22 ). The list of lipid genes with NCBI identifiers is shown in online Supplementary Table S1. RNA-Seq analysis was performed using R (version 3.4.1). The KEGG ontology enrichment analysis (KOEA) and DEA were conducted using R package edgeR. DEA was conducted using pairwise exact tests to produce gene fold changes and P values. Genes with a false discovery rate-adjusted P value (q) <0·05 were considered differentially expressed between two test conditions. Hypergeometric test was applied in KOEA, based on the number of differential expressed genes (DEG) v. total genes annotated to each KEGG ontology (KO) term, and differences were considered significant when P<0·05. A heatmap was drawn using R with the pheatmap package. The pathway diagram was produced using PathVisio version 3.2.4. All other figures were produced using R package ggplot2.
Results
Intestinal fatty acid composition
The percentage of major fatty acids in PtdCho, PtdEtn and TAG of salmon after feeding PO compared with FO is shown in Table 2. Other fatty acids are shown in online Supplementary Table S2. Regardless of tissue, fatty acid composition of TAG was similar to the fed diet (Table 2). The content of 18 : 1n-9, 18 : 2n-6 and 18 : 3n-3 increased after feeding PO compared with FO, whereas the content of 20 : 1n-9, 22 : 1n-11, EPA and DHA decreased. The number of significantly (P<0·05) changed fatty acids was much higher in fingerlings (2·5 g) than in juveniles (10 g), and the changes in percentage of fatty acids were also higher. For example, the percentage of DHA in TAG was reduced by over 50 % in PO compared with FO-fed fingerlings, whereas in juveniles the TAG-DHA was reduced by 30 %.
Table 2 Fatty acid composition of phosphatidylcholine (PtdCho), phosphatidylethanolamine (PtdEtn), and TAG in stomach, pyloric caeca and hindgut of fingerlings (2·5 g) and juvenile (10 g) salmonFootnote †(Mean values and standard deviations)
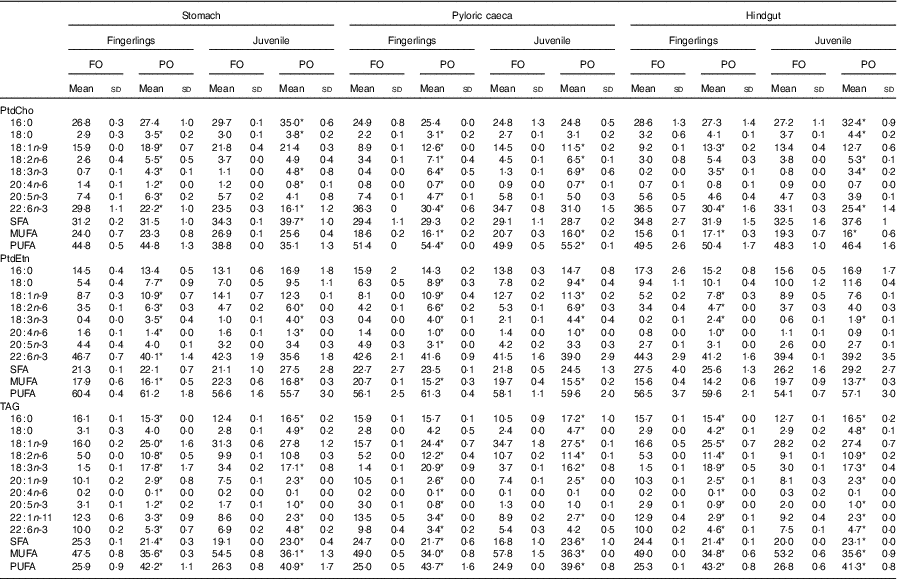
FO, fish oil; PO, plant oil.
* Significantly (P<0·05) different fatty acid composition in PO group compared with FO group.
† Two biological and technical replicates (2×2) were used in the analysis.
The dominant fatty acids of PtdCho were 16 : 0, 18 : 1n-9, EPA and DHA, and feeding of PO had a different impact on the PtdCho of different intestinal regions and developmental stages of salmon (Table 2). The amount of 16 : 0 was relatively stable regardless of diet, except for a slight increase in stomach and hindgut of juvenile salmon fed PO. The content of 18 : 1n-9 increased in all three tissues of fingerlings after feeding PO, but in juveniles the effects were less pronounced in pyloric caeca, with no effects observed on stomach and hindgut. The percentage of 18 : 2n-6 and 18 : 3n-3 increased in both fingerling and juvenile salmon fed PO compared with FO, which was in line with the high amount of the two fatty acids in the PO diet (Tables 1 and 2). Reductions of ARA and EPA were only observed in stomach and pyloric caeca of fingerlings after feeding PO, whereas DHA decreased in all three tissues. The EPA and DHA levels were not affected in pyloric caeca of juvenile salmon when fed PO diet.
The composition of fatty acids in PtdEtn was less influenced by PO diet compared with the fatty acids in other lipid classes (Table 2). The content of 16 : 0 was not changed when fed PO, whereas a slight increase in 18 : 0 was observed. The percentage of 18 : 1n-9 was only increased in fingerlings, similarly observed in PtdCho. Increases in 18 : 2n-6 and 18 : 3n-3 were also found when feeding PO compared with FO. The content of EPA was significantly (P<0·05) reduced in pyloric caeca of salmon fingerlings; otherwise, no difference was observed between PO and FO groups. Over 40 % of DHA were found in PtdEtn of all intestinal regions, and the percentage was only reduced in stomach of fingerlings when fed PO.
Overview of intestinal transcriptome and KEGG ontology analysis
An average of twenty-two million reads were collected from each library, of which approximately 85 % were mapped to the salmon genome. From a total of 81 597 genes currently annotated, 35 381 genes met a minimum level of expression for DEA (at least 1 CPM in two or more samples). The highest number of DEG between PO and FO treatments was 2214 found in pyloric caeca of fingerlings, which reduced to 486 DEG in juveniles. The number of DEG in stomach also decreased from 437 in fingerlings to six in juveniles. On the other hand, the DEG in the hindgut and liver of fingerlings were 142 and 40, respectively, which both increased to approximately 500 DEG in juveniles.
The highest number of significantly (P<0·05) regulated KEGG pathways was found in pyloric caeca of fingerlings (Fig. 1). These included ten pathways involved in lipid metabolism, five pathways in carbohydrate metabolism and six pathways in protein metabolism. Fewer KEGG pathways were significantly (P<0·05) regulated in pyloric caeca of juvenile salmon, including only three pathways involved in lipid metabolism. The pathway of steroid biosynthesis was highly regulated both in fingerling and juvenile salmon fed PO compared with those fed FO. Feeding of PO also caused significant (P<0·05) changes of three pathways of fatty acid metabolism in pyloric caeca of fingerlings, indicating a group of DEG involved in fatty acid metabolism. Pathways of glycerolipid and glycerophospholipid metabolism were also changed in pyloric caeca of fingerlings fed PO compared with FO. Many pathways were changed in stomach of fingerlings fed PO diet, whereas few pathways were changed in the same tissue of juvenile salmon. In the hindgut, three pathways of lipid metabolism were changed in fingerlings after feeding PO, whereas no lipid metabolism pathway was changed in juveniles. However, thirteen pathways in carbohydrate and protein metabolism were significantly regulated in juvenile salmon fed PO. No lipid metabolism pathway was regulated in liver when feeding PO diet. All other significantly (P<0·05) regulated pathways in stomach, pyloric caeca, hindgut and liver are shown in online Supplementary Table S3.
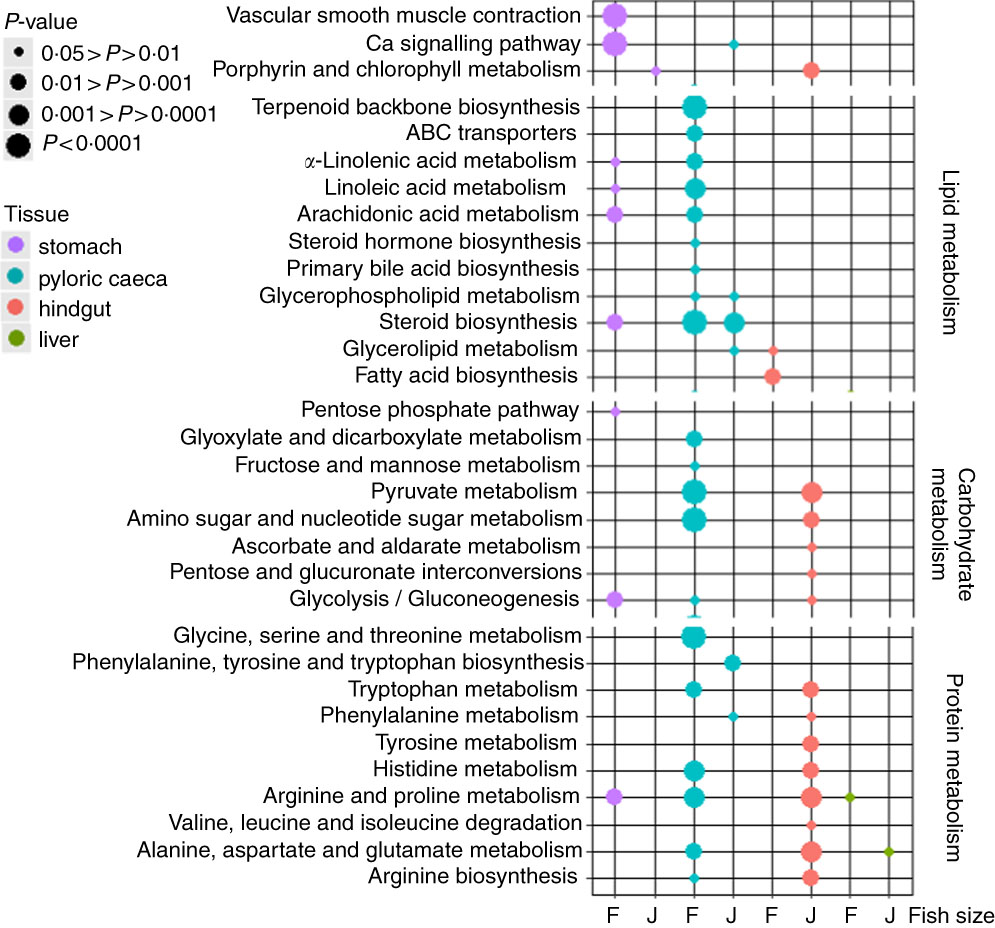
Fig. 1 Bubble graph of KEGG ontology enrichment analysis results for stomach, pyloric caeca, hindgut and liver of fingerling (F, 2·5 g) and juvenile (J, 10 g) salmon. Various KEGG pathways involved in lipid, carbohydrate and protein metabolism were significantly (P<0·05) regulated after feeding plant oil (PO) compared with fish oil. Statistical test was applied using hypergeometric test based on the number of PO-induced differential expressed genes v. total genes annotated to each KEGG ontology term.
Regulatory divergence of Ss4R gene duplicates
Out of 8147 identified Ss4R duplicate pairs in total, about 5500 pairs had similar expression (log2 fold change between −0·5 and 0·5) in any tissue of salmon fed PO compared with FO. The rest of the pairs contained at least one DEG (log2 fold change >0·5 or <−0·5) after feeding PO, including approximately 400 duplicate pairs with both increased or decreased expression (log2 fold change both >0·05 or <−0·05), and around 100 pairs were reversely expressed. More interestingly, the sum and difference of fold change in expression of the two duplicate genes were larger in fingerlings than in juveniles (Fig. 2). This difference of regulation between fingerlings and juveniles was more distinct in intestinal tissues rather than in the liver. The total number of correlated and reversely expressed Ss4R duplicate pairs induced by PO was also reduced in intestinal tissues, whereas it increased in liver from fingerling to juvenile salmon (online Supplementary Table S4).
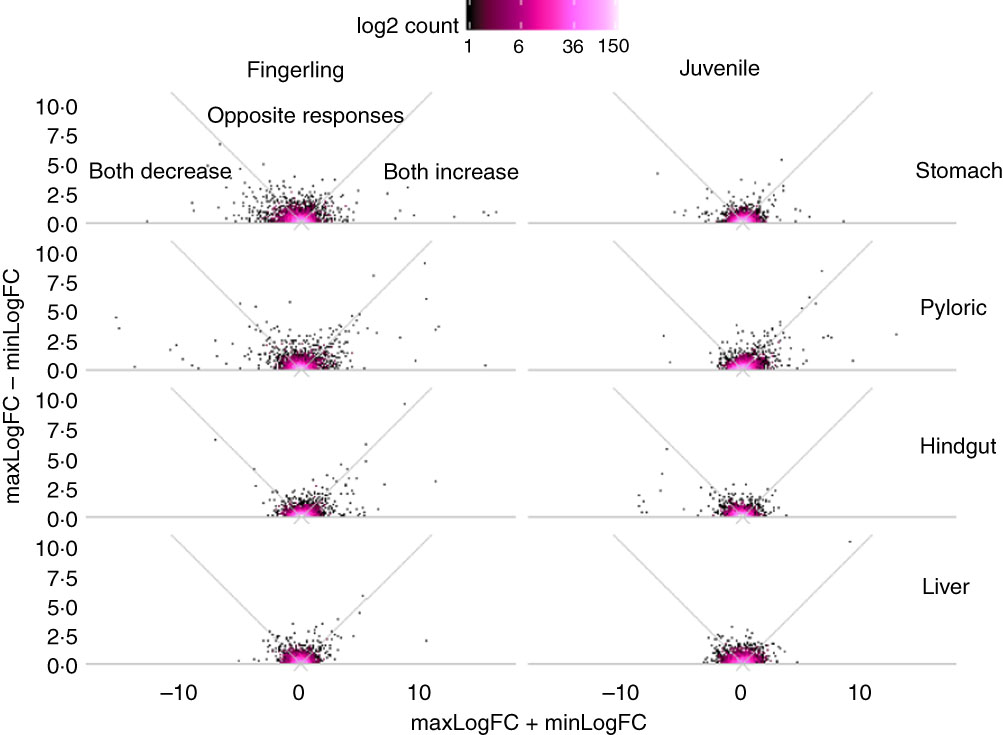
Fig. 2 Plant oil (PO) induced expressional change of salmonid-specific (Ss4R) duplicate genes in fingerling (2·5 g) and juvenile (10 g) salmon. Each duplicate pair was separated into the gene with higher fold change (maxLogFC) and lower fold change (minLogFC) after feeding PO, where maxLogFC>minLogFC. The regulation divergence of Ss4R duplicate pairs can be separated into three groups: both increased (maxLogFC>0, minLogFC>0), both decreased (maxLogFC<0, minLogFC<0) and opposite response. ![]() , Accumulation of duplicate pairs within each dot area.
, Accumulation of duplicate pairs within each dot area.
Expressional differences of lipid metabolism genes among tissues
Most of the 344 genes for lipid metabolism were highly expressed in pyloric caeca or liver, whereas few genes were highest expressed in stomach or hindgut (Fig. 3). In pyloric caeca, most lipid metabolism genes showed increasing expression from fingerling to juvenile stages, whereas very few genes had decreasing expression. Genes with increasing expression during development were found in hindgut and liver, but this pattern of change was mostly found in cholesterol and fatty acid metabolism pathways. Regardless of fish size, key genes in pathways of LC-PUFA elongation and desaturation, cholesterol biosynthesis and transport and lipoprotein formation were highly expressed only in pyloric caeca or liver (online Supplementary Table S5). These genes were also expressed in the hindgut, but at a lower level. The expression of these pathways was very low in the stomach. However, some genes encoding for fatty acid binding proteins were highly expressed in stomach and hindgut. Genes for fatty acid elongase elovl1b_1 and elovl7ba were also highly expressed in stomach, whereas other elongase and desaturase genes had very low expression levels.
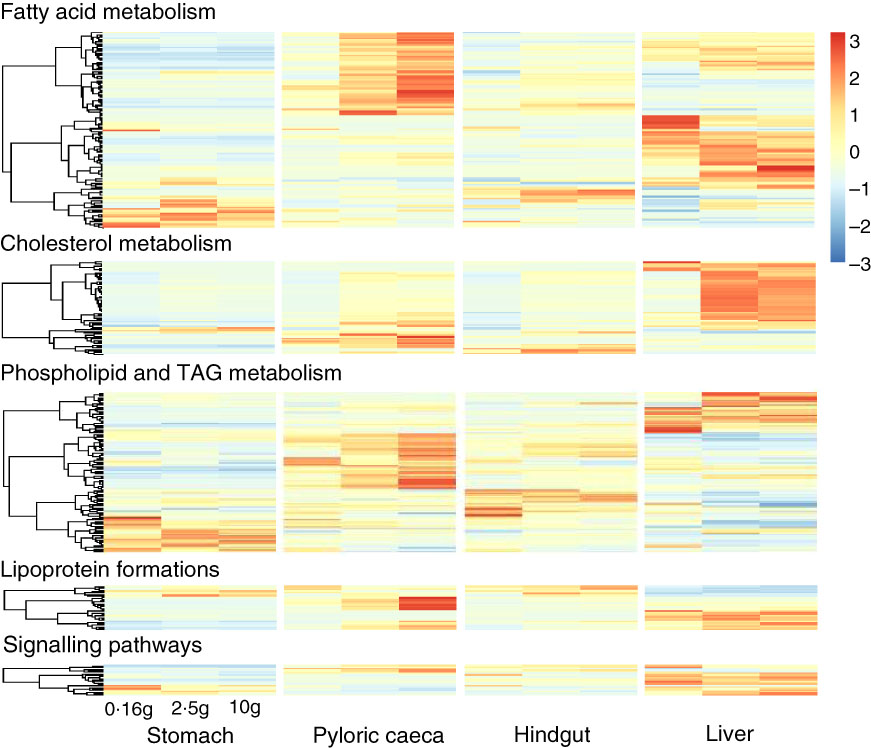
Fig. 3 Heatmap of 344 genes for lipid metabolism between different tissues. Different clusters of genes were dominant in each tissue and developmental stage of salmon. For each tissue, the three columns represent average transcripts per million (TPM) for samples of 0·16, 2·5 and 10 g salmon from left to right. The colour intensity is relative to the standard deviation from mean of TPM over developmental stages and tissues (row-scaled).
Regulation in lipid metabolism pathways in different tissues
A total of seventy-six DEG were identified in pyloric caeca of fingerlings, whereas only eleven DEG were found in juveniles (online Supplementary Table S6). The DEG in fingerlings included twenty-six DEG covering each enzymatic step of the pathway for cholesterol biosynthesis (Fig. 4(a)). The expression of these genes was higher in fingerlings fed PO compared with those fed FO, indicating up-regulated cholesterol biosynthesis. However, these PO-induced differences of expression were evened out in juveniles and only five DEG were found. This was because of increased expression of genes for cholesterol biosynthesis in FO-fed juveniles compared with fingerlings, whereas the gene expression in PO-fed salmon was relatively unchanged between the two developmental stages.
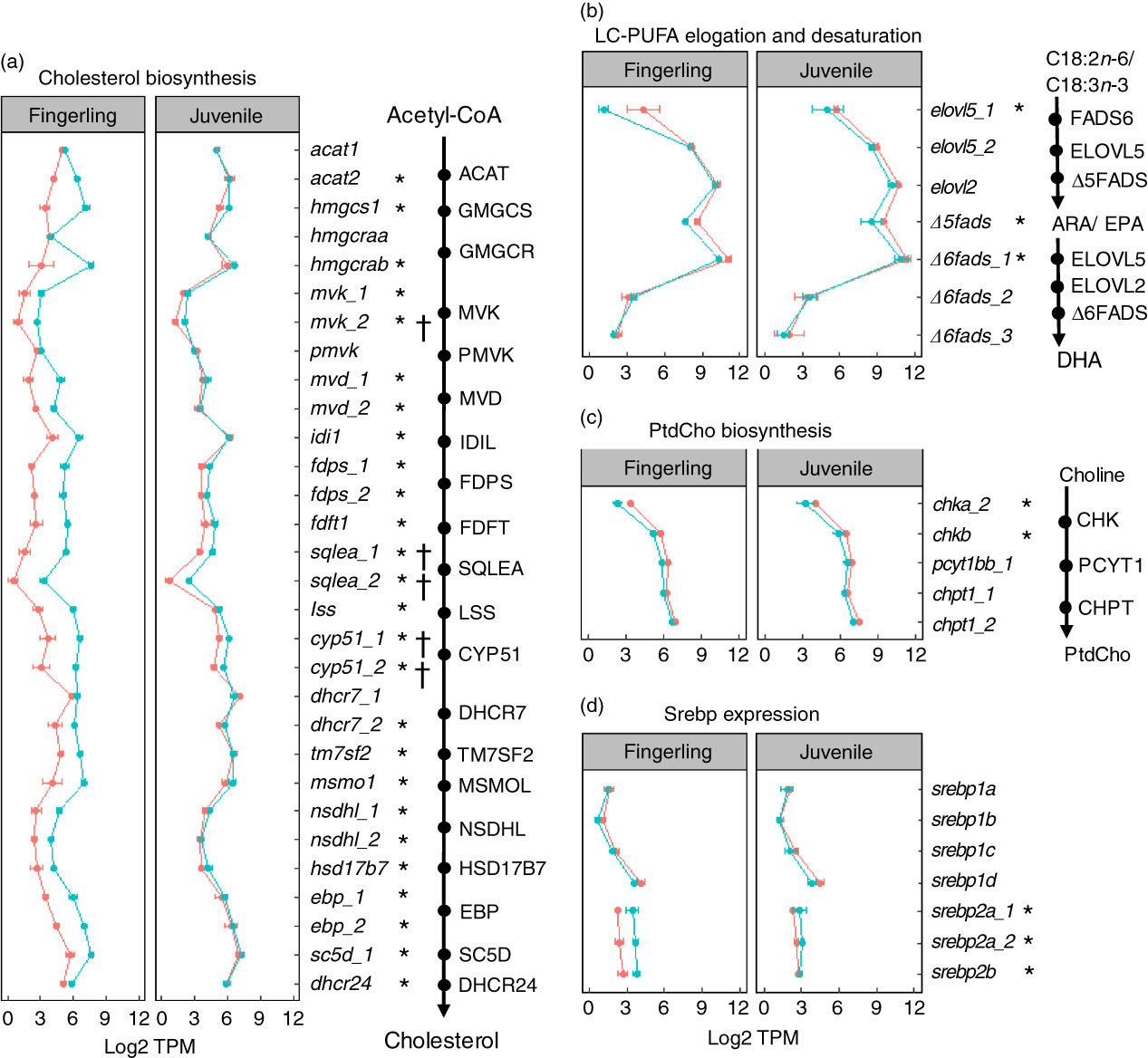
Fig. 4 Genes for lipid metabolism in pyloric caeca of salmon fingerlings (2·5 g) and juveniles (10 g) fed either fish oil (FO, ![]() ) or plant oil (PO,
) or plant oil (PO, ![]() ). The expression of genes is compared in transcripts per million (TPM). * or †, Differentially expressed gene (q<0·05) in fingerling or juvenile salmon, respectively, fed PO compared with FO. (a) Expression of genes involved in each enzymatic step in the pathway of cholesterol biosynthesis. Gene duplicates are shown in a few steps of the pathway, which encode enzymes controlling the same chemical reaction. Higher expression of genes is shown for the PO group of fingerlings than for the FO group, whereas such differences are less clear in juvenile salmon. (b) Expression of seven key genes involved in long-chain PUFA (LC-PUFA) elongation and desaturation pathway. Expression was throughout slightly higher in salmon fed FO than PO diet. (c) Expression of five key genes involved in biosynthesis of phosphatidylcholine (PtdCho), which is the major phospholipid in the pyloric caeca of salmon. The expression of genes was higher in salmon fed FO diet than those fed PO diet. (d) Expression of seven srebp gene duplicates that play an important role in regulating genes in cholesterol and fatty acid biosynthesis in pyloric caeca of salmon. The srebp1 genes were more highly expressed in FO-fed salmon, whereas srebp2 genes were more highly expressed in PO-fed salmon.
). The expression of genes is compared in transcripts per million (TPM). * or †, Differentially expressed gene (q<0·05) in fingerling or juvenile salmon, respectively, fed PO compared with FO. (a) Expression of genes involved in each enzymatic step in the pathway of cholesterol biosynthesis. Gene duplicates are shown in a few steps of the pathway, which encode enzymes controlling the same chemical reaction. Higher expression of genes is shown for the PO group of fingerlings than for the FO group, whereas such differences are less clear in juvenile salmon. (b) Expression of seven key genes involved in long-chain PUFA (LC-PUFA) elongation and desaturation pathway. Expression was throughout slightly higher in salmon fed FO than PO diet. (c) Expression of five key genes involved in biosynthesis of phosphatidylcholine (PtdCho), which is the major phospholipid in the pyloric caeca of salmon. The expression of genes was higher in salmon fed FO diet than those fed PO diet. (d) Expression of seven srebp gene duplicates that play an important role in regulating genes in cholesterol and fatty acid biosynthesis in pyloric caeca of salmon. The srebp1 genes were more highly expressed in FO-fed salmon, whereas srebp2 genes were more highly expressed in PO-fed salmon.
On the other hand, the pathway of fatty acid elongation and desaturation was down-regulated in pyloric caeca after feeding PO compared with FO (Fig. 4(b)). Key elongase gene elovl5_1 was strongly down-regulated (log2 fold change=−4·3, q=7·6 × 10−8) in fingerling salmon fed PO. Two desaturase genes Δ5fads and Δ6fads_1, with Δ5 and Δ6 activities, respectively, were also highly expressed (q<0·05) in FO-fed salmon compared with PO. Although no DEG was found in the pathway of elongation and desaturation in juvenile salmon, the expression of elovl5_1, Δ5fads and Δ6fads_1 was still higher in FO-compared with PO-fed salmon.
The expression of genes in phospholipid biosynthesis was mostly down-regulated in pyloric caeca of salmon fed PO (Fig. 4(c)). The chkb gene, which is involved in the first step of PtdCho biosynthesis( Reference Vance and Vance 35 ), was significantly (q<0·05) higher expressed in salmon fingerlings fed FO compared with those fed PO. Genes in the subsequent two steps of PtdCho biosynthesis, pyct1bb_1 and chpt1_2, were also more highly expressed in FO than in PO. Although the expression of genes in PtdEtn biosynthesis was less clear than the gene in PtdCho pathways, slightly elevated expression of genes was found in FO-fed salmon (online Supplementary Table S6). Genes for sterol regulatory element-binding protein (SREBP) were regulated differently between duplicates (Fig. 4(d)). Genes srebp2a_1, srebp2a_2 and srebp2b were all significantly (q<0·05) down-regulated in salmon fingerlings after feeding PO compared with FO, whereas the srebp1 genes srebp1b and srebp1d were up-regulated. The expressional differences between the two dietary groups were smaller in juvenile salmon compared with fingerlings, and no DEG was found.
Many genes involved in lipid absorption and transport were differentially expressed in pyloric caeca of fingerling salmon, whereas almost no DEG was found in juveniles (online Supplementary Table S6). The absorption of cholesterol was increased after feeding PO compared with FO, as two key genes scarb1 and npc1l1 were up-regulated in pyloric caeca of fingerlings (Fig. 5). Genes abca1ab_1 and abca1ab_2, which were involved in excretion of cholesterol into circulation( Reference Oram and Vaughan 36 ), were also highly expressed in PO-fed salmon. The absorption and intercellular transport of fatty acids in pyloric caeca were largely influenced by feeding PO. Most fatty acid transporter genes, namely cd36_2, fabp2b, fabp6_1 and fatp4_2, were more highly expressed in pyloric caeca of salmon fingerlings fed PO than FO, whereas fatp2c was more lowly expressed. The expression of cd36_2 and fabp6_1 was still higher in salmon juvenile fed PO compared with FO. The pathway of lipoprotein formation in pyloric caeca was more lowly expressed in PO-fed salmon as key genes such as mtp_1, apoa1_1 and sar1ba_1 were down-regulated.
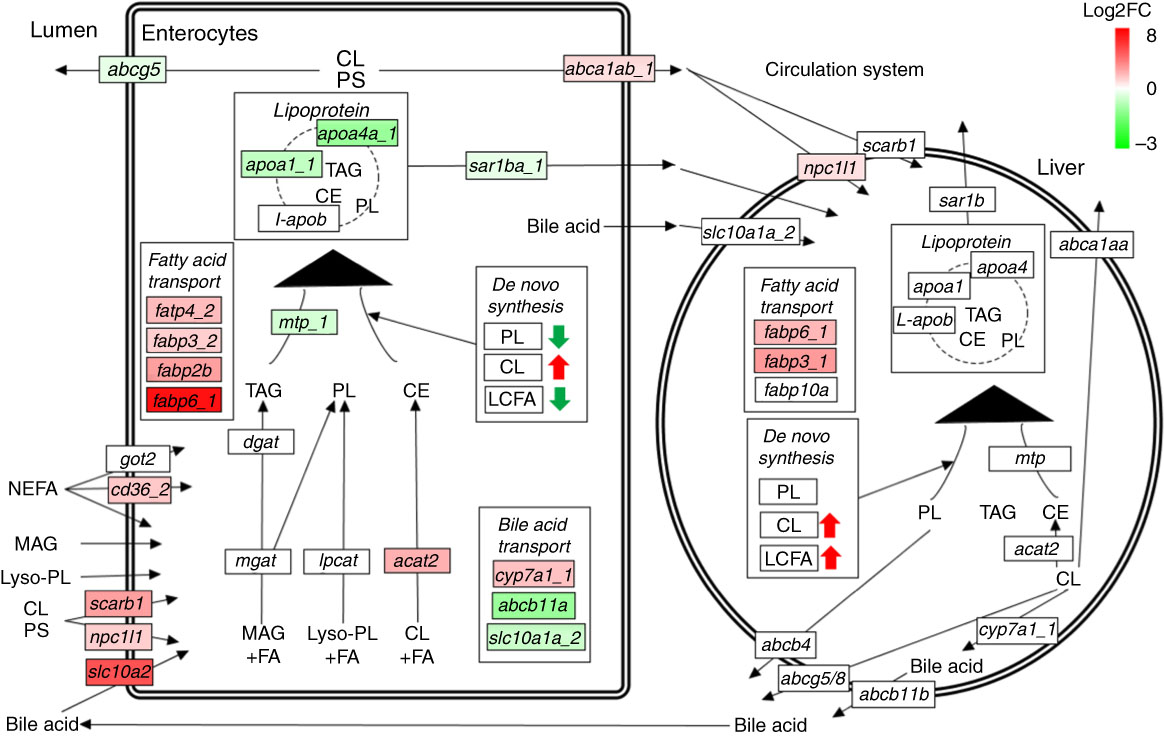
Fig. 5 Pathways of lipid metabolism in intestine and liver of salmon. Dietary NEFA, monoacylglycerol (MAG), lyso-phospholipids (lyso-PL), cholesterol (CL) and phytosterol (PS) are absorbed by intestinal enterocytes through either diffusion or a group of transport proteins. MAG, lyso-PL and CL are resynthesised into TAG, phospholipids (PL) and cholesteryl ester (CE), respectively, which are then packed into lipoprotein and effused to the circulatory system. Absorbed CL and PS can also be directly exported back to intestinal lumen or circulatory system through different transport proteins. Liver can take up lipoprotein residues and other nutrients from the circulatory system, and re-package them into new lipoproteins for circulation. Both intestine and liver have abilities of synthesising TAG, PL, CL and LCFA de novo. The labels indicate the highest expressed gene encoding the proteins. Only significantly expressed genes (q<0·05) are marked in colour. The colour intensity is relative to the log2 fold change (Log2FC) of genes in plant oil-fed salmon compared with fish oil-fed salmon.
No clear size effect was found in liver on expressional differences of genes for lipid metabolism between fingerling and juvenile salmon (online Supplementary Table S6). The genes for cholesterol biosynthesis and fatty acid elongation and desaturation were all more highly expressed in salmon fed the PO diet compared with those fed the FO diet, although no DEG was identified. Two fatty acid transporter genes fabp6_1 and fabp3_1 and one cholesterol transporter gene npc1l1 were significantly (q<0·05) more highly expressed in PO-fed salmon rather than FO (Fig. 5). This indicates up-regulated pathways of cholesterol biosynthesis and fatty acid elongation and desaturation in liver after feeding PO. Few DEG were found to be involved in the pathways of lipid metabolism in stomach and hindgut of salmon after feeding PO (online Supplementary Table S6). Fatty acid binding protein genes fabp10a_1 and fabp10a_2 were significantly (q<0·05) up-regulated in stomach after feeding PO, whereas fabp3_1 and fabp2b were up-regulated in the hindgut.
Discussion
The present study aimed to increase current knowledge on how dietary PO affects lipid metabolism in developing salmon fingerlings (2·5 g) and juveniles (10 g). PO diets contained lower levels of LC-PUFA and cholesterol, which resulted in many biochemical and genetic feedback responses related to lipid metabolism in stomach, pyloric caeca, hindgut and liver. Many DEG were involved in cholesterol and fatty acid metabolism in pyloric caeca of fingerlings, with high fold changes of expression after feeding PO compared with FO. Both the number of DEG and their level of expression in PO- and FO-fed groups were much higher in fingerlings than in juveniles. This suggests that lipid metabolism regulation in fingerlings is more responsive to a plant-based diet than in juveniles. There were no DEG in liver related to lipid metabolism pathways in either fingerling or juvenile salmon after feeding PO. This contrasts a recent study on pre-smolt (approximately 50 g) and post-smolt (approximately 200 g) salmon fed the same PO and FO diets used in this present study, where a much higher number of lipid metabolism DEG were found in liver compared with intestine( Reference Gillard, Harvey and Gjuvsland 22 ). Therefore, we suggest that fingerling and early juvenile salmon have an immature liver with low activity of lipid metabolism, which gradually increases as the fish develops.
De novo cholesterol synthesis was the most influenced pathway found in pyloric caeca of fingerlings when fed PO, with DEG covering every enzymatic step of the pathway. This is probably related to the reduced dietary cholesterol in fish fed PO, inducing increased cholesterol de novo synthesis( Reference Siperstein 37 ). However, the requirement of cholesterol is probably lower in juveniles than in fingerlings, suggested by the up-regulation of cholesterol biosynthesis genes in fingerlings fed PO, while the expression of these genes remained similar in juveniles. A slight up-regulation of cholesterol biosynthesis was also found in liver, suggesting that liver was partly responsible for synthesising cholesterol to compensate for a dietary shortage. Besides de novo synthesis, fingerlings can also regulate their body cholesterol level through increasing the uptake of cholesterol while reducing its efflux. We found that the scarb1 and npc1l1 genes, which are responsible for cholesterol uptake( Reference Jia, Betters and Yu 38 ), and the abca1ab_1 gene, which is responsible for transporting cholesterol into circulation( Reference Oram and Vaughan 36 ), were up-regulated in pyloric caeca of fingerlings after feeding PO( Reference Fumeron, Bard and Lecerf 39 ). On the other hand, the abcg5 gene, which is responsible for ejecting cholesterol back the intestinal lumen( Reference Field, Born and Mathur 40 ), was down-regulated. The increased biosynthesis and absorption of cholesterol can be regulated by SREBP, which are transcription factors controlling cellular cholesterol content( Reference Brown and Goldstein 41 ). Two groups of SREBP have been identified in mammals, from which SREPB-2 is more selective for genes involved in cholesterol homoeostasis( Reference Horton, Shimomura and Brown 42 , Reference Pai, Guryev and Brown 43 ). The higher expression of srebp2 genes in pyloric caeca of PO-fed salmon was in line with the up-regulation of cholesterol biosynthesis genes, in agreement with a previous finding in seabream( Reference Houston, Karalazos and Tinsley 44 ).
Dietary PO often contains high amounts of phytosterols, which can compete with cholesterols for space in the micelles and reduce cholesterol absorption in the intestine( Reference Liland, Espe and Rosenlund 20 , Reference Amemiya-Kudo, Shimano and Hasty 45 ). Owing to its similarity to cholesterol, phytosterols can be taken up in the intestine and transported to fish body tissues by the same transport proteins used for cholesterol( Reference Liland, Espe and Rosenlund 20 , Reference Fumeron, Bard and Lecerf 39 ). However, phytosterols do not appear to be esterified by acyl-CoA:cholesterol O-acyltransferase 2 (ATAC2) and packed into lipoproteins with TAG and phospholipids( Reference Temel, Gebre and Parks 46 ). Rather, phytosterols seem to mainly be exported back into the intestinal lumen through ATP-binding cassette transporter sub-family G (ABCG5/8), whereas the rest can be transported into circulation through ATP-binding cassette transporter sub-family A (ABCA1)( Reference Fumeron, Bard and Lecerf 39 , Reference Field, Born and Mathur 40 ). In fingerlings, the increase in expression of abca1ab_1, encoding ABCA1, and the decrease in expression abcg5, encoding ABCG5, could increase the accumulation of phytosterols in the intestinal tissue, and consequently transport into circulation. Previous studies have shown that dietary phytosterols did not influence growth and performance of parr or adult salmon( Reference Sissener, Liland and Holen 23 , Reference Gu, Kortner and Penn 47 ). However, whether phytosterol has an impact on fingerling or juvenile salmon needs to be further investigated.
In general, the rate of changes in fatty acid composition after feeding PO compared with FO was consistently larger in fingerlings than in juveniles. This suggests that salmon fingerlings have less metabolic regulatory control when fed the PO diet than juveniles. Regarding the lipid classes, the fatty acid composition of TAG was often similar to the composition of dietary fatty acids, whereas phospholipids, which are the membrane lipids and thus play structural roles, are often more strictly regulated( Reference Sargent, Tocher and Bell 48 ). However, feeding of PO had a major effect on most fatty acids in both PtdCho and PtdEtn. This suggests that PtdCho and PtdEtn can use a wide range of diacylglycerol with different fatty acid compositions for their carbon skeleton( Reference Olsen, Dragnes and Myklebust 17 ). Dietary PO had particularly little effect on the percentage of PtdCho-DHA and almost no effect on PtdEtn-DHA, suggesting that DHA was selectively retained in phospholipid when salmon were fed PO( Reference Stubhaug, Lie and Torstensen 49 ). Similar to previous findings in adult trout( Reference Tarnawski, Hollander and Stachura 50 ), a higher percentage of 18 : 1n-9 and ARA was more efficiently retained in stomach than in pyloric caeca and hindgut. The high percentage of ARA was probably used for eicosanoid synthesis and gastric mucosa protection( Reference Tarnawski, Hollander and Stachura 50 ).
Although dietary PO increased the 18 : 3n-3 and 18 : 2n-6 contents and decreased ARA, EPA and DHA percentages in pyloric caeca of fingerlings, the expression of elongation (elovl5_1) and desaturation (Δ5fads and Δ6fads) genes was depressed. This was associated with down-regulation of srebp1 genes, which are the primary transcription factors for endogenous production of LC-PUFA in salmon( Reference Carmona-Antonanzas, Tocher and Martinez-Rubio 51 ). These results are in disagreement with the generally accepted conclusion that dietary PO stimulates the expression of genes for elongation and desaturation in fish( Reference Tocher 15 , Reference Kjaer, Ruyter and Berge 52 ). The reason for the down-regulation of elongation and desaturation in pyloric caeca of PO-fed fingerlings remains unclear. A similar result has been found using primary cultures of enterocytes obtained from fish fed linseed oil( Reference Tocher, Fonseca-Madrigal and Bell 16 ), which was also used in this study. The activity of elongation and desaturation in enterocytes was significantly reduced in PO compared with FO fed salmon, whereas in hepatocytes the activity was increased( Reference Tocher, Fonseca-Madrigal and Bell 16 ). We also identified slight up-regulation of elovl5, elovl2, Δ5fads and Δ6fads genes in liver following PO feeding. The reason for the reverse regulation of elongation and desaturation between pyloric caeca and liver was not clear.
For the first time in salmon, we identified a variety of genes involved in fatty acid transport pathways and measured their expression among intestinal and liver tissues. Pyloric caeca are the predominant regions for nutrient and fatty acid uptake in fish. This explained the higher expression of fatty acid transporter cd36 and fatp genes in the pyloric caeca than in other intestinal tissues. These genes had lower, but considerable, expression in hindgut, in line with a lower but still notable fatty acid uptake in salmonids( Reference Olsen, Myklebust and Kaino 4 , Reference Olsen, Dragnes and Myklebust 17 ). Unlike mammalian CD36 mRNA expressions( Reference Chen, Yang and Braunstein 53 , Reference Wilson, Tran and Erion 54 ), salmon cd36_1 and cd36_2 genes had low expression levels in liver. However, we found that fatp genes had higher expression in liver than intestinal tissues, suggesting that it is primarily responsible for fatty acid uptake in the tissue. Dietary PO stimulated varieties of fatp and cd36 genes in pyloric caeca tissues, indicating increased activity of fatty acid uptake and transport. On the other hand, fatp2c was down-regulated in pyloric caeca, probably owing to the different fatty acid preference by fatty acid transporters. A previous study has demonstrated that dietary rapeseed oil with high 18 : 1n-9 could reduce cd36 and fatp expression in salmon( Reference Torstensen, Nanton and Olsvik 55 ). Furthermore, the intracellular transport of fatty acids in liver and intestine can be largely affected by dietary PO, as different fabp genes were differentially expressed in stomach, pyloric caeca, hindgut and liver.
Bile acids are synthesised from cholesterol by cholesterol 7α-hydroxylase (CYP7A1) in liver and transported through transporter ATP-binding cassette, sub-family B member 11 (ABCB11) into the gallbladder where they are mixed with phospholipids, then transported through ABCB4 to form mature bile. In the intestine, bile acids can be reabsorbed through the SLC10A2 protein and returned to portal circulation before being taken up by liver through SCL10A1 protein and repumped into the gallbladder( Reference Trauner and Boyer 56 ). In mammals, the cyp7a1, abcb11, abcb4 and scl10a1 genes are highly expressed in liver (available on https://www.proteinatlas.org/). We also found high expression levels of cyp7a1, abcb11, abcb4 and scl10a1 genes in pyloric caeca being differentially expressed in fingerlings fed PO compared with those fed FO. This could suggest an additional bile acid production and transport in pyloric caeca, which could be especially important for salmon at early developmental stages. However, the function and activity of the proteins encoded by these genes are largely unknown in salmon, and it remains unclear whether these proteins have similar functions as in liver.
Whole-genome duplication is believed to play a major role in evolution, providing extra copies of genes with sub- or neo-functionalisation, on which selection can take place( Reference Wolfe 57 ). This study showed that 23 % (350 out of 1512) of the salmon duplicated pairs were reversely regulated after feeding PO compared with FO, indicating a high regulatory divergence of duplicates in salmon. The incomplete rediploidisation of the salmon genome has increased the complexity of the pathways of lipid metabolism in the species. It has been suggested that salmon duplicates may have led to diverged gene expressions between different tissues such as intestine and liver( Reference Lien, Koop and Sandve 32 ). However, as the intestine consists of different regions with distinct functions, it is physiologically and genetically important to analyse the functional divergence of these regions separately. The regulation of salmon duplicates was different between stomach, pyloric caeca and hindgut. This is in accordance with the physiological similarity between the tissues, where pyloric caeca is responsible for digestion and absorption, whereas stomach is more responsible for pepsin and acid degradation and physical decomposition of the feed.
In conclusion, the present study has provided a systemic overview of PO-induced changes in lipid metabolism in intestine and liver of salmon during early stages of development. By comparing the expression levels of lipid metabolism genes between PO and FO, we found that salmon has a more sensitive response to PO at fingerling stages compared with juvenile. In particular, the expression levels of genes in cholesterol absorption and biosynthesis and transport pathways in pyloric caeca were highly up-regulated in fingerlings, suggesting a higher requirement of dietary cholesterol. An increased cholesterol biosynthesis pathway was also found in liver, although no clear difference was found between fingerling and juvenile stages. The fatty acid elongation and desaturation pathways were down-regulated in pyloric caeca of fingerlings after feeding PO diet, but up-regulated in liver. However, further in vivo or in vitro studies are required to confirm the present findings on protein level. We also suggest future studies on dietary requirement of lipids in fingerling stages of salmon, as the fish seemed to have insufficient capacity of lipid metabolism at early stages of development.
Acknowledgements
The authors thank Hanne Hellerud Hansen, Thomas Nelson Harvey and the Centre for Integrative Genetics for RNA-Seq sample preparation and data analysis. The authors thank Dr Maren Mommens and AquaGen AS for providing experimental facilities and other practical information, and Dr Keshuai Li for support on lipid analysis.
The design, analysis and writing of this article was supported by no specific grant from Department of Biology, Norwegian University of Science and Technology (NTNU). The RNA-Seq and data analysis were financed by the Research Council of Norway (GenoSysFat, grant no. 244164), (DigiSal, grant no. 248792). The authors thank AquaGen AS for providing the fish and for contribution of running the experiment. The feeds were produced by Dominic Nanton and Cargill Innovation Centre Dirdal. The authors also thank the China Scholarship Council for providing financial support to Yang Jin for his PhD study.
Y. J., Y. O. and R. E. O. designed the research; Y. J. and G. B. G. performed the RNAseq and transcriptomic data analysis; Y. J. performed the lipid analysis; J. O. V. and S. R. S. guided the transcriptomic analysis and revised the manuscript. Y. O. and R. E. O. guided the lipid analysis and revised the manuscript. M. Ø., N. S. and S. A. K. provided input on the experimental design, carried out the feeding trial and sampling and reviewed the manuscript. All authors read and approved the final manuscript.
The authors declare that there are no conflicts of interest.
Supplementary material
For supplementary material/s referred to in this article, please visit https://doi.org/10.1017/S0007114518001885










