INTRODUCTION
Vibrio cholerae, a Gram-negative γ-proteobacteria, is the causative agent of cholera disease and is acquired through the ingestion of contaminated food or surface water. Globally 1·3 billion cases of acute diarrhoea occur in children aged <5 years annually [Reference Sur1]. Only two serogroups namely, O1 and O139 are recognized as pathogenic and responsible for epidemic and pandemic cholera [Reference Kaper2]. Similarly non-O1/non-O139 serogroups of V. cholerae may cause sporadic diarrhoeal episodes with mild to severe morbidity [Reference Kaper2–Reference Ramamurthy4].
Studies on phages of V. cholerae have been of historical interest. Among several typing methods, phage typing is one of the most important and useful methods for the identification and differentiation of V. cholerae strains. A phage-typing scheme was developed by Basu & Mukerjee for V. cholerae O1 biotype El Tor [Reference Basu and Mukerjee5] and later a new phage-typing scheme was developed for O1 [Reference Chattopadhyay6]. Subsequently, a further phage-typing scheme specific for V. cholerae O139 was developed at the National Institute of Cholera and Enteric Diseases (NICED) [Reference Chakraborty7]. As a WHO collaborating centre for diarrhoeal diseases research and training, NICED is a national reference laboratory and receives annually between 700 and 1000 strains of V. cholerae from different parts of India and abroad for biotyping, serotyping and phage typing [Reference Sarkar8]. Phage-typing methods can test large numbers of strains rapidly. It is a cost-effective and a simple laboratory method that does not require any sophisticated equipment. Because of host specificity it offers the basic information on identification, biotyping and discrimination of strains [Reference Bhowmick9].
Increasingly, laboratory diagnosis, identification and epidemiological surveillance of cholera are based on genotypic characteristics rather than phenotypic characteristics (biochemical and antigenic properties), including antibiotic-resistance testing, biotyping, serotyping and phage typing. Conventional typing methods can not accurately predict the epidemiological potential of V. cholerae strains [Reference Stroeher10] due to low discriminatory power. The use of genome-based bacterial identification and typing would reveal an epidemic relationship and the association of certain clones with potential sources of contamination [Reference Barrett11–Reference Vadivelu15]. Several DNA-based methods, e.g. detection of virulence genes, pulsed-field gel electrophoresis (PFGE) and random amplification of polymorphic DNA (RAPD) are now extensively used for typing of V. cholerae strains. Following the advent of DNA-based strain typing methods, there is need for a suitable strain typing method that must be rapid, cost-effective and with high discriminatory power.
Variable number of tandem repeats (VNTR) has been demonstrated to be superior in its fine-tuning discriminatory power for some enteric pathogens. This method is also applied in V. cholerae genomes by PCR amplification and size determination of simple sequence repeats and DNA sequencing of mononucleotide repeat regions identified in the V. cholerae genomes [Reference Danin-Poleg16–Reference Rosche, Yano and Oliver18]. These molecular loci themselves became known as VNTR regions and are now well-established targets for ancestry analysis [Reference Jeffreys19]. The existence of slipped-strand mispairing as a basic molecular mechanism of DNA variation in VNTR regions is generally accepted [Reference Levinson and Gutman20]. The repeat number indicates the Mendelian inheritance, may vary within a species, and represents a unique genetic feature of an individual [Reference Vodopyanov21]. The availability of the full genome sequence of V. cholerae enables computerized analysis of the presence of repetitive DNA with VNTR-like potential. In this study, the VcA locus has been selected as it has already been employed in identifying various strains of the cholera pathogen. We evaluated the potential of VcA VNTR analysis as a tool for genotyping of V. cholerae.
METHODS
Bacteriology
A total of 125 V. cholerae isolates were included in this study. These strains were collected from different geographical regions of India and abroad in different time-frames and sent to NICED for confirmation, biotyping, serotyping and phage typing (Table 1). Sixty-seven strains of O1 El Tor, 29 of V. cholerae O139, 23 non-O1/non-O139 and six O1 classical strains were included. V. cholerae O1 biotype El Tor strains MAK 757 (ATCC 51352) and N16961, V. cholerae O1 biotype classical strains CL-154 and O395 and V. cholerae O139 strain NPR-4 were used as controls [Reference Basu and Mukerjee5, Reference Chattopadhyay6, Reference Basu22, Reference Chakrabarti, Ghosh and Sarkar23]. All these strains were maintained in nutrient agar stabs and kept in the dark, at the Vibrio Phage Reference Laboratory, NICED. All strains of V. cholerae were identified and confirmed by using standard techniques [24]. A single colony of V. cholerae from nutrient agar was inoculated into Mueller–Hinton broth and incubated under stationary conditions for 4 h at 37°C. The cultures were then plated on Mueller–Hinton agar plates containing 50 μg/ml (122·34 USP unit/ml) of polymyxin B sulphate (Sigma, USA) and incubated at 37°C overnight [Reference Basu22]. V. cholerae O1, O395 and MAK 757 were used as negative and positive controls, respectively. These strains were subsequently serotyped by using polyvalent O1, monospecific Ogawa and Inaba antisera (Difco, USA). This study is a continuation of our previously published work where 26 strains (PL-1 to PL-26) were analysed by antibiogram, phage typing, PFGE and RAPD–PCR [Reference Bhowmick9].
Table 1. Summarized data on year of isolation, geographical locations, phage types, PFGE types, RAPD types and VcA VNTR types of V. cholerae strains incorporated in this study
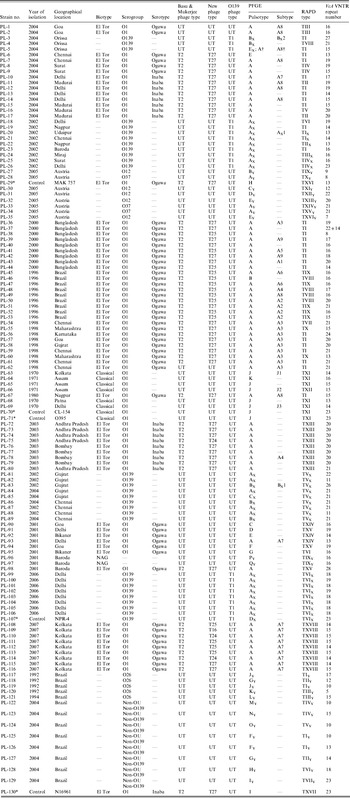
UT, Untypable.
* These strains were used as control strains in this study.
† This V. cholerae O139 strain had a similar PFGE type as the El Tor strain.
Antimicrobial susceptibility testing
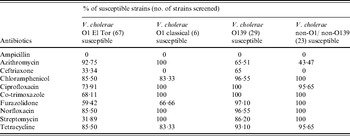
Antimicrobial susceptibility testing was performed by the disk diffusion method with commercially available disks (HiMedia, India). All the V. cholerae strains were examined for resistance to ampicillin (10 mg), azithromycin (15 mg), ceftriaxone (30 mg), chloramphenicol (30 mg), ciprofloxacin (5 mg), co-trimoxazole (25 mg), furazolidone (100 mg), norfloxacin (10 mg), streptomycin (10 mg) and tetracycline (30 mg) according to criteria of the National Committee for Clinical Laboratory Standards [25]. Strains with intermediate zone of inhibition were interpreted as resistant [Reference Yamamoto26]. The American Type Culture Collection (ATCC) strain E. coli (ATCC 25922) was used as a standard quality control.
Phage-typing procedure
The phage typing was performed by the standard methodology of our laboratory [Reference Chattopadhyay6, Reference Chakrabarti, Ghosh and Sarkar23, Reference Sarkar27]. MAK 757 (ATCC 51352) and NPR-4 [Reference Chakrabarti, Ghosh and Sarkar23] were used as control for O1 and O139 strains, respectively. Additionally, ϕ149 of classical group IV and El Tor group V phage were used to differentiate the biotype of V. cholerae strains.
RAPD
A modified method from the study of Murray & Thompson [Reference Murray and Thompson28] was used for DNA extraction. RAPD–PCR fingerprinting was carried out with 25 ml of reaction mixture containing 2·5 ml of 10 μl PCR buffer [500 mm KCl, 100 mm Tris–HCl (pH 8·0), 15 mm MgCl2 and 0·01% gelatin], 20 ng genomic DNA, 20 pmol primer 1281 (5′-AACGCGCAAC-3′) [Reference Akopyanz29], 1 U AmpliTaq DNA polymerase and 2·5 ml of 25 mm dNTP for 45 cycles of 94°C for 1 min, 36°C for 1 min and 72°C for 2 min in a Peltier Thermal Cycler (PTC-200; MJ Research, USA). After PCR, 10 ml of product were electrophoresed in 2% agarose gel containing 0·5 mg/ml ethidium bromide and photographed under UV light using the Gel Doc 2000 documentation system (Bio-Rad, USA). The 1 kb DNA ladder (Sigma, USA) was used as the molecular size standard.
PFGE
Genomic DNA was prepared in agarose plugs as described previously [Reference Kurazono30]. NotI [5′-GCGGCCGC-3′] (Takara, Japan) digested inserts of V. cholerae were applied to a contour-clamped homogenous electric field in a CHEF Mapper System (Bio-Rad) using 1% PFGE grade agarose in 0·5× Tris-borate-EDTA buffer for 40 h 24 min at 14°C. Run conditions were generated by autoalgorithm mode of CHEF Mapper that separates the DNA size range of 20–300 kb. After electrophoresis, the gel was stained in distilled water containing 1·0 mg ethidium bromide/ml for 30 min, distained in distilled water for 15 min and photographed under UV light using the Gel Doc 2000 documentation system (Bio-Rad). A DNA size standard (PFGE ladder; New England Biolabs, USA) was used as the molecular size standard.
PFGE gel images were retrieved and aligned to generate a composite image containing the banding profiles of all the isolates. The image was analysed with Quantity One 1-D Analysis Software (version 4.4.0, Bio-Rad) to detect the relatedness of isolates. Bands ranging from 34·4 to 543·2 kb were considered for the construction of a dendrogram. Pattern profiles were compared by using Dice coefficient and UPGMA (unweighted pair-group method with arithmetic mean) clustering with a 4% tolerance window. A dendrogram showing the hierarchical representation of the level of linkage between the isolates was drawn to predict the degree of clonal relatedness.
VcA VNTR
DNA extraction of pure cultures was prepared by a method modified from the work of Murray & Thompson [Reference Murray and Thompson28]. For crude and fast DNA extraction a suspension of 109 cells in distilled waster was heated in boiling water for 15 min, then immediately chilled on ice for 10 min and centrifuged at 10 000 rpm. The supernatant was immediately used for amplification. To amplify a DNA fragment containing VcA, two specific primers, 5′-TCT TCT TGC GCT TCT TGA CC-3′ (direct) and 5′-TCA TCA AGA TGC ACG ACA CA-3′ (reverse) were used [Reference Vodopyanov21]. The amplification product containing 23 repeated units was expected to be 222 nt in size in the control strain N16961. PCR mixtures contained 2·5 μl of 10× PCR buffer [500 mm KCl, 100 mm Tris–HCl (pH 8·0), 15 mm MgCl2 and 0·01% gelatin], 50 ng of template DNA, 10 μm concentrations of each primer, 2·5 μl of 2·5 mm dNTP and 0·2 U of AmpliTaq DNA polymerase (Takara) in a total volume of 25 μl. The reaction was carried out in a Peltier Thermal Cycler (PTC-200; MJ Research) as follows: 94°C for 5 min; followed by 30 cycles of 1·5 min at 94°C, 1·5 min at 50°C and 1·5 min at 72°C. All PCR amplification products were analysed by agarose gel (1·5%) electrophoresis.
PCR products were purified using a QIAquick PCR purification kit (Qiagen, USA). Portions (50 ng) of the purified product were sequenced on both strands using a BigDye terminator cycle sequencing kit (Applied Biosystems, USA) and loaded onto an ABI 3100 Genetic Analyzer sequencer (Applied Biosystems) according to the manufacturer's instructions. Analysis of the results was done by using the sequence analysis software Lasergen software suit (DNAStar Inc., USA).
Nucleotide sequences of VcA regions of all the V. cholerae strains incorporated in this study were analysed with DNADIST 3·5c to detect the relatedness of the isolates. This program can use a neighbour phylogenetic tree interface to compute distance matrix from nucleotide sequences. Sequence profiles were compared by this program using the neighbour-joining method. A dendrogram showing the hierarchical representation of the level of linkage between the isolates was drawn to predict the degree of clonal relatedness.
RESULTS
Antibiotic sensitivity profile
All the O1 El Tor and O139 isolates were uniformly resistant to ampicillin (Table 2). All O1 classical and non-O1/non-O139 strains were resistant to ampicillin and ceftriaxone. In total, 68·1% V. cholerae O1 El Tor isolates were resistant to streptomycin, whereas a higher level of susceptibility was observed in O139, classical and non-O1/non-O139 isolates against this antibiotic. In the current study, all the V. cholerae strains were not uniformly susceptible to tetracycline. Higher susceptibility was seen in all strains except non-O1/non-O139 tested with azithromycin. All strains showed high susceptibility against ciprofloxacin, norfloxacin, chloramphenicol, co-trimoxazole, furazolidone, ciprofloxacin and chloramphenicol.
Table 2. Antimicrobial susceptibility pattern of V. cholerae strains incorporated in this study
Phage-typing results
Phage-typing results are given in Table 1. During the study period all the O1 El Tor strains were clustered into phage type T2 with Basu & Mukerjee's scheme. It was evident from Table 2 that type T27 was the predominant type (56·7%). The phage types other than type T27 did not follow any regular pattern and appeared scattered in different parts of the world. Except for type T27 other phage types were type T25 (22·3%), type T24 (4·4%), type T16 (1·4%) and type T4 (1·4%). Out of 67 strains, 12·1% were untypable with the new typing scheme. The remaining 29 O139 isolates (68·9%) from different geographical regions of India were grouped as type T1 and 31·0% were untypable with the O139 typing scheme. All O1 classical and non-O1/non-O139 strains were untypable with both the O1 and O139 typing scheme.
RAPD analysis
Purified genomic DNA isolated from V. cholerae isolates was used for DNA fingerprinting by the RAPD method. The results were obtained with primer 1281 (5′-AACGCGCAAC-3′) with amplified fragment size ranging from 0·2 kb to 10·0 kb. RAPD–PCR was used as a means to determine the clonal relatedness of isolates by their chromosomal polymorphism. Using RAPD–PCR out of 130 V. cholerae strains 69 O1 El Tor strains gave rise to 16 different electrophoretic patterns TI–TX and TXIII–TXVIII (Table 1). RAPD type TI was the most prevalent, and includes 33·3% isolates belonging to both serotypes of Ogawa and Inaba. This was followed by TXIII (13·0%), TXVIII (13·0%), TIX (7·2%) and TVIII (5·7%). Eight V. cholerae O1 classical strains were grouped into two types TXI (10·1%) and TXII (1·4%) and that were different from O1 El Tor strains. Only six distinct RAPD types TIX (6·6%), TIIX (3·3%), TIIIX (3·3%), TIVX (6·6%), TVX (30%) and TVIX (30%) were found in 30 V. cholerae O139 strains. Six of the O139 strains were presented as TI (four isolates), TVI (one isolate) and TVII (one isolate) which shared 100% similarity with these RAPD types of O1 El Tor strains. Much more heterogeneity was observed within 23 V. cholerae non-O1/non-O139 strains, i.e. from TIY to TXVY. RAPD patterns obtained with reference V. cholerae strains MAK 757 (PL-29, TXVI), N16961 (PL-130, TXVII), CL-154 (PL-70, TXI), O395 (PL-71, TXI) and NPR-4 (PL-107, TVIX) were different from each other except CL-154 and O395, while all these strains were dissimilar in their RAPD profile from the rest of the strains included in this study.
Analysis of restriction digestion patterns of PFGE and cluster analysis
To assess the genetic interrelationship of V. cholerae strains, 130 V. cholerae strains were analysed by PFGE using the restriction endonuclease NotI. PFGE patterns were classified basically as proposed by Tenover et al. [Reference Tenover, Arbeit and Goering31]. PFGE of all strains exhibited fragments ranging in size from 34·4 kb to 543·2 kb. A total of 33 pulsotypes were identified in 130 strains typable by PFGE (Table 1). Of the 33 distinctive pulsotypes, nine pulsotypes were represented by V. cholerae O1 El Tor (A–I). Pulsotype A was the predominant pulsotype, consisting of 59 O1 El Tor strains and showed nine subtypes (A1–A9) which were closely related to each other. The remaining pulsotypes (B–I) of O1 El Tor strains were distinct from pulsotype A and from each other. Two different PFGE patterns (pulsotypes J and K) were noted in the V. cholerae O1 classical strains and pulsotype J comprised of nine O1 classical strains exhibited three subtypes (J1–J3). The PFGE patterns of O1 El Tor and O1 classical were totally different from each other. Of 29 V. cholerae O139 strains, 22 appeared to have same banding pattern, pulsotype AX and this pulsotype showed one subtype AX1 consisting of one O139 strain. The other pulsotypes detected in O139 strains were BX (five isolates) with two subtypes BX1 (one isolate) and BX2 (one isolate), CX (one isolate), DX (one isolate, i.e. control strain NPR-4) and EX (one isolate). Interestingly, the EX pulsotype of O139 serogroup gave a similar PFGE pattern as the A8 subtype of V. cholerae O1. By PFGE, all non-O1/non-O139 isolates were genetically very diverse, as 17 different patterns (pulsotypes AY–QY) were identified in 22 typable isolates by PFGE. The PFGE profiles of non-O1/non-O139 strains differed from each other and from the patterns displayed by the strains belonging to the O1 and O139 serogroups. Furthermore, PFGE patterns A8 and A4 predominated among Ogawa serotypes whereas subtype A was predominant within Inaba serotypes.
A dendrogram of 130 V. cholerae strains was constructed of nine major well separated clusters (Supplementary Fig. S1, available online). The O1 El Tor, O1 classical, O139 and non-O1/non-O139 strains were constructed separately from their PFGE profiles. For O1 El Tor strains, the four major clusters were detected and they were closely related. The strains of these clusters were PL-2, PL-8, PL-12, PL-15, PL-77, PL-78; PL-7, PL-9, PL-11, PL-16, PL-80, PL-79, PL-76, PL-116, PL-115, PL-112; PL-114, PL-108, PL-113, PL-111, PL-109, and PL-62, PL-61, PL-60, PL-57, PL-56, PL-55 and PL-54. V. cholerae O139 strains revealed two major clusters, one comprised of PL-18, PL-19, PL-21, PL-22, PL-23, PL-25 and PL-26 strains and these strains showed closely similar PFGE profiles with the strains PL-20 and PL-81. The second major O139 cluster with strains PL-99 and PL-101 to PL-106 showed 89% similarity with the first cluster of O139. The O1 classical and non-O1/nonO139 V. cholerae strains were in separate clusters.
VcA loci distribution within V. cholerae genome
Amplification with the two specific primers for the VcA locus revealed high VcA polymorphism in all V. cholerae strains incorporated in this study. In total, 22 types of VcA alleles with a minimum of 5 to a maximum of 28 repeat numbers were found in the V. cholerae strains examined; the locus was detected in every strain (Table 1). The TGCTGT repeat number was established for each allele. Allele sizes in all samples were estimated against a molecular-weight marker (Fig. 1) and the results were verified by direct sequencing of the allele found in 130 V. cholerae strains of this study. Nucleotide sequences of the 3′ terminal of the VcA region of all the strains are given in Supplementary Figure S2 (online). The repeat number 16 was the predominant type containing 17 strains (11 O1 El Tor, one O1 classical, three O139 and two non-O1/non-O139). Repeat number 15 was observed in 16 strains, 21 in 14 strains, 13 in eight strains, 19 in nine strains, 14 in 12 strains, 20 in 16 strains, 23 in six strains, eight in two strains, 22 in two strains, 18 in nine strains and 12 in two strains. The three VcA repeats, repeat 28 in one strain, repeat 17 in four strains and repeat 24 in one strain were detected only in V. cholerae O1 El Tor strain. Out of the 130 V. cholerae strains incorporated in this study, only one (PL-37) represented a cell mixture with two VcA alleles, 22 and 14. The VcA repeats with 11 (two strains), 26 (one strain) and 28 (one strain) repeat numbers were detected only in four V. cholerae O139 strains but were totally absent in all other O1 El Tor, classical and non-O1/non-O139 strains. Subsequently, repeat number 9 (one strain), number 4 (four strains) and number 5 (one strain) were observed only in non-O1/non-O139 strains. Dendogram analysis of 130 V. cholerae isolates (Supplementary Fig. S3, online) showed 11 major well separated clusters with repeats 16, 15, 14, 23, 13, 21, 17, 18, 19, 20 and 10. The V. cholerae O139 strains from different parts of India appeared as a separate branch of the dendrogram with 89% of the bands being common.
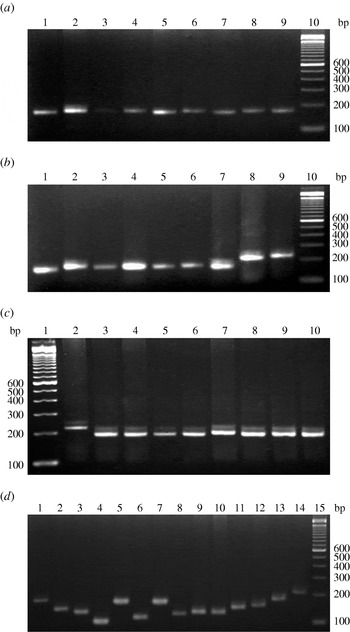
Fig. 1. VcA VNTR profiles of representative V. cholerae strains of this study. (a) Representative V. cholerae O1 El Tor strains: lane 10, 100-bp DNA ladder; lanes 1–9 contain strains PL-108 to PL-116, respectively. (b) V. cholerae O1 classical strains: lane 10, 100-bp DNA ladder; lanes 1-4 contain strains PL-63 to PL-66, lane 5 contains O1 El Tor strain PL-67 and lanes 6–9 contain strains PL-68 to PL-71, respectively. (c) Representative V. cholerae O139 strains: lane 1, 100 bp DNA ladder; lanes 2–10 contain strains PL-99 to PL-107, respectively. (d) Representative V. cholerae non-O1/non-O139 strains: lane 15, 100 bp DNA ladder; lanes 1–14 contain strains PL-117 to PL-130, respectively.
Comparative study of phenotypic and genotypic methods
Of the 130 V. cholerae strains, seven sets of O1 El Tor strains PL-1, PL-2; PL-45, PL-47; PL-57, PL-58; PL-59, PL-61, PL-62; PL-72, PL-73, PL-74, PL-76, PL-77, PL-79; PL-108, PL-112 and PL-11, PL-113, one set of O1 classical PL-70, PL-71 and three sets of V. cholerae O139 strains PL-82, PL-88; PL-99, PL-101, PL-102, PL-103, PL-104, PL-105, PL-106 and PL-100, PL-102 which belonged to similar serogroups also gave similar phage type, RAPD type, PFGE profile and same number of repeats in VcA VNTR. Under O1 El Tor strains type T27 was the predominant type and included 41 V. cholerae strains but showed 12 PFGE types (four pulsotypes and eight subtypes), 12 RAPD types and 13 types of VcA alleles. The second major phage type T25 consisting of 15 isolates was differentiated into eight PFGE types (two subtypes and six pulsotypes), three RAPD types and seven types of VcA repeats. Three O1 El Tor strains with similar phage type T24 were dissimilar in their PFGE (two types), RAPD (two types) and VcA VNTR (three repeats) results. A single strain PL-109 was identical in its PFGE (subtype A7), RAPD (TVIII) profile and VcA repeat number [Reference Vadivelu15] with O1 El Tor strains but showed totally different phage type, i.e. T16. Eight V. cholerae O1 El Tor strains were untypable with both the O1 typing scheme but molecular-typing methods were able to type them and gave seven types of PFGE (five pulsotypes and two subtypes), three types of RAPD patterns and four types of VcA repeats. On the other hand, the eight O1 classical V. cholerae isolates exhibited entirely different PFGE (two pulsotypes and three subtypes) and RAPD (TXI and TXII) profile from the rest of the strains in this study but with similar VcA repeats. Within V. cholerae O139 serogroup 21 strains with phage type T1 were discriminated by their PFGE (four pulsotypes and one subtype), RAPD (five types) and VcA VNTR (nine types of VcA alleles) profile. Similar results (PFGE, three pulsotypes; RAPD, one type; and VcA repeat, four types) were also seen in the nine untypable O139 strains. Interestingly, five O139 strains PL-3, PL-4, PL-18, PL-19 and PL-23 gave RAPD profiles such as TI, TVIII and TVI similar to O1 El Tor strains. Subsequently, another O139 strain PL-5, exhibited both O1 El Tor PFGE (subtype A8) and RAPD (TI) profiles. In this context 23 V. cholerae non-O1/non-O139 strains demonstrated 17 entirely different PFGE pulsotypes and 15 different RAPD types from the remaining V. cholerae strains of this study. They produced 15 types of VcA VNTR alleles and among them only repeat numbers 5, 10, 12 were found. In the current study, we have also found a geographical relationship between strains of V. cholerae. Some sets of strains such as PL-1, PL-2; PL-45, PL-47; PL-59, PL-61, PL-62; PL-76, PL-77, PL-79; PL-72, PL-73, PL-74, PL-75 originated in Goa, Brazil, Chennai, Bombay, and Andhra Pradesh, respectively and were indistinguishable from each other by all the molecular-typing tools. Therefore, these sets of strains were genetically and also geographically related. Interestingly, two large sets of strains PL-99 to PL-106 and PL-108 to PL-116 originated from the same geographical location were genetically similar by PFGE and RAPD but distinguishable by VcA VNTR.
DISCUSSION
The intent of this study was to discover a swift, cost-effective and more discriminatory molecular genotyping tool of V. cholerae strains that would facilitate the continuous monitoring and extensive epidemiological analysis of cholera outbreaks. For this purpose different molecular-typing methods, RAPD, PFGE and VcA VNTR, were used to understand the genetic relatedness between the V. cholerae strains isolated from different geographical regions, within the country and abroad, that had different phage types.
An antibiogram of V. cholerae strains revealed that 68·1% V. cholerae O1 El Tor isolates were resistant to streptomycin, whereas a higher level of susceptibility was seen in O139, classical and non-O1/non-O139 isolates with this drug. Therefore, it appears that one drug, streptomycin, could allow differentiation between the strains belonging to serotypes O1 El Tor, O139 and non-O1/non-O139 of V. cholerae. As previously reported, there was no significant correlation between phage types and antibiogram [Reference Sarkar8].
Phage typing is a conventional typing system, which has been used for decades at NICED, India. Because of its high host specificity it offers basic information on identification, biotyping and discrimination of strains. Theoretically, strains belonging to a single phage type originate from a single clone. However, in the current study, a high level of heterogeneity was found in RAPD, PFGE, VcA VNTR and computer-based analysis of genomic profiles which indicated the clonal diversity of V. cholerae O1 and O139 serogroups. In parallel to phage typing, molecular-typing methods should be included for efficient integrated national and international epidemiological research on V. cholerae.
Molecular-typing techniques have the potential for studying the origin and interrelatedness of strains of epidemiological settings [Reference Sur1]. PFGE, RAPD–PCR and VcA VNTR showed that V. cholerae strains belonging to same phage type were genetically heterogeneous except for a few sets of strains. Interestingly, five O139 strains gave RAPD profiles such as TI, TVIII and TVI similar to O1 El Tor strains and another O139 strain PL-5, exhibited both O1 El Tor PFGE (subtype A8) and RAPD (TI) profiles. This indicates that the V. cholerae O139 probably originated from O1 ancestry. The clustering of isolates can vary considerably between RAPD and PFGE. The RAPD method does not enable the constitution of a pattern for the discrimination of V. cholerae O1 strains. Furthermore, the reproducibility of the reaction conditions poses a problem for routine laboratory use. This inconvenience makes it complicated to create a schematic pattern for use in public health laboratories to identify the different clones for epidemiological investigation.
On the contrary, PFGE has proven to be the best technique for investigating the clonal relatedness amongst enteric pathogens, e.g. E. coli O157:H7, Campylobacter jejuni, Vibrio fluvialis, Listeria monocytogenes, Clostridium perfringens, Shigella dysenteriae, etc. [Reference Stroeher10, 32–Reference Pazhani36]. In our previous study, PFGE was more discriminatory than phage typing as the predominant phage type was divided into multiple pulsotypes [Reference Bhowmick9]. PFGE had better typability and concordance with different geographical regions than phage typing as reported earlier from our laboratory [Reference Bhowmick9].
VNTRs are DNA regions consisting of several tandemly repeated oligonucleotides and represent a distinctive hereditary feature of an individual. VNTR has been used by some workers as a strain typing tool for V. cholerae by exploiting several VNTR loci [Reference Danin-Poleg16, Reference Stine37, Reference Ghosh38]. VcA VNTR was used in the current study to type the V. cholerae strains. Computer analysis revealed seven potential VNTR loci in the DNA sequences of V. cholerae chromosomes 1 and 2 [Reference Vodopyanov21]. One of these VcA, was chosen in this study. This locus was selected for further analysis by Vodopyanov et al. [Reference Vodopyanov21], because it has the highest repeat number and thereby allows a greater allele polymorphism in various strains. The fact of being chosen established VcA VNTR as a most informative genetic typing tool for V. cholerae [Reference Vodopyanov21]. However, phylogenetic analysis revealed the clustering of V. cholerae strains from different parts of India and abroad, indicating a clonal relationship. Dendogram analysis of VcA VNTR produced 11 well separated clusters with clonal relatedness of V. cholerae strains from different geographical regions of India and abroad. VcA VNTR detects all genetic variations within the amplified VcA fragment while PFGE examines only those that are in specific restriction sites and those that involve large insertions or deletions of DNA. As variation occurs most commonly at the nucleotide level, it could be suggested that the discriminatory ability of VcA VNTR is better than PFGE. This fact was also strongly reflected in our results. In particular, results obtained from two batches of strains PL-99 to PL-106 and PL-108 to PL-116 clearly showed the typing abilities of VcA VNTR and PFGE. VcA VNTR analysis is a rapid, cost-effective, and easy to implement typing method with high discriminatory power. This method also combines genome-wide coverage of loci with different mutation rates, which could serve as an efficient tool for phylogenetic studies and for rapid bacterial typing. The results obtained from our study also exhibited and corroborated this fact and validate VcA VNTR as an important genetic typing tool for V. cholerae.
NOTE
Supplementary material accompanies this paper on the Journal's website (http://journals.cambridge.org/hyg).
DECLARATION OF INTEREST
T.S.B. and M.D. were Senior Research Fellows of the National Institute of Cholera & Enteric Diseases and B.L.S was Deputy Director of NICED at the time of the study. The authors and NICED have no financial interests that are affected by the material in the paper.





