Recently, strong associations between variants in the human Δ-5 and Δ-6 desaturase genes fatty acid desaturase 1 (FADS1) and fatty acid desaturase 2 (FADS2) on chromosome 11q12-11q13.1 and fatty acid composition in serum phospholipids have been detected by our group(Reference Schaeffer, Gohlke, Muller, Heid, Palmer, Kompauer, Demmelmair, Illig, Koletzko and Heinrich1). In particular, the all-minor haplotype (which is composed of the minor allele at each locus) reconstructed from five loci (rs174544, rs174553, rs174556, rs174561, rs3834458) showed very strong associations with the n-6 fatty acid arachidonic acid (20 : 4n-6), which often functions as a precursor of inflammatory eicosanoids (4-series leucotrienes and 2-series prostaglandins), depending on the metabolic situation. Further strong associations of the all-minor haplotype were observed with other n-6 fatty acids (18 : 2n-6, 18 : 3n-6, 20 : 2n-6 and 20 : 3n-6) and to a lesser extent with the n-3 fatty acids 18 : 3n-3, 20 : 5n-3 and 22 : 5n-3. These results are important as it has been shown in several studies that this region on chromosome 11q12-11q13.1 is linked with atopy(Reference Cookson, Sharp, Faux and Hopkin2–Reference Wheatley, Bolland, Hewitt, Dewar and Hall6). In fact, in our previous study, it was shown that carriers of the rare alleles of several single nucleotide polymorphisms (SNP) and their respective haplotypes had a lower prevalence of allergic rhinitis and atopic eczema. As such associations may be mediated by products of the arachidonic acid pathway, however, it is crucial to demonstrate that these haplotypes are also associated with the substrates, i.e. membrane PUFA.
In the present study we aimed (i) to replicate the reconstructed haplotypes and their associations with fatty acids in blood phospholipids as observed in the previously studied population of the European Community Respiratory Health Survey (ECRHS) in Erfurt, East Germany, using an independent population sample(Reference Hoff, Seiler and Heinrich7). In addition we (ii) wanted to test for effects of these haplotypes at the cellular level, using erythrocyte membranes as proxy membranes. The latter is an interesting study question, since the proof of an effect would have implications for other cell types with no or low desaturase activities, including leucocyte sub-populations.
Materials and methods
Study design and population
The Bavarian Nutrition Survey II (BVS-II) was designed as a representative study of the Bavarian population to investigate dietary and lifestyle habits(Reference Hoff, Seiler and Heinrich7). Between September 2002 and June 2003, about 1000 subjects of the German-speaking population in Bavaria (Germany) aged 13–80 years were recruited by a three-stage sampling procedure. At baseline, the subjects' lifestyle and socio-economic characteristics as well as their health status were assessed by means of a computer-aided personal interview. The overall participation rate in the study was 71 % (n 1050). In the following weeks, the participants were contacted three times by phone in order to collect information on dietary intake and physical activity (24 h recalls). Within 6 weeks after recruitment, adults (age ≥ 18 years) who completed at least one 24 h dietary recall (n 879) were invited to their nearest health office for blood sampling and standardised anthropometric measurements. Of these, 65 % (n 568) provided blood samples, with one participant denying the use of DNA for further studies. Information on SNP and fatty acid composition of erythrocyte membranes as well as plasma phospholipids and SNP data were available for 535 and 163 participants, respectively. Note that the latter 163 participants are a subset of the 535 participants of the BVS-II study for whom PUFA was determined both in plasma and in erythrocytes. Only marginal differences by age, socio-economic status or smoking status were observed between study participants invited for blood sampling and those finally providing blood samples. All participants gave their written informed consent. The study was approved by the local ethics committee.
Blood sampling
Venous blood was drawn into EDTA tubes (1·6 mg/l blood), chilled at 4°C, and processed subsequently. Plasma and buffy coat (leucocytes) were separated from erythrocytes by centrifugation at 2000 g for 15 min. Samples were cooled for a maximum of 1 d (transportation, sampling) until they were stored at − 80°C (plasma) or − 24°C (erythrocytes). The analysis of erythrocyte membrane fatty acids was conducted no later than 6 months after drawing.
Fatty acid analysis
A sample of 0·5 ml erythrocyte suspension was used for membrane fatty acid analysis. The erythrocyte membranes were isolated by centrifugation at 20 000 g for 20 min at 4°C and the pellet was suspended with TRIS buffer (11 mm-2-amino-2-hydroxymethyl-propane-1,3-diol, 1 mm-NaEDTA, pH 7·4). The washing procedure was repeated twice before adding 800 μl distilled water(Reference Golik, Weissgarten, Evans, Cohen, Averbukh, Zaidenstein, Cotariu and Modai8). Lipid extraction from erythrocyte membranes and plasma (1 ml) was performed with chloroform–methanol (2/1, v/v) according to a modification of the method of Folch et al. (Reference Folch, Lees and Sloane Stanley9). Briefly, lipids were extracted twice with a chloroform–methanol mixture, containing the antioxidant butylated hydroxytoluene (50 mg/l). The combined extracts were washed with a CaCl2 solution. The organic phase was collected and evaporated to dryness. Phospholipids were isolated from other plasma lipids by means of TLC. The dried extracts were re-suspended in chloroform and fatty acid methyl esters were obtained by transesterification with trimethylsulfonium hydroxide(Reference Butte10).
Fatty acid methyl esters were separated using a 100 m CP-Sil-88 capillary column (Varian-Chrompack, Darmstadt, Germany), installed in an HP 5890 series II gas chromatograph with a flame-ionisation detector (Hewlett Packard, Munich, Germany). For identification and quantification of the fatty acid methyl ester peaks, authentic standards were used (Sigma Aldrich, Steinheim, Germany). In total, the content of twenty-two types of fatty acids were determined and expressed as a percentage of the total fatty acids identified; for each sample the mean result of two injections is given. By means of a 6-fold analysis of pooled erythrocytes, CV for the most prominent fatty acids (> 1 % of all fatty acid methyl esters) were between 0·4 and 5·1 %.
Genotyping
Calculating the linkage disequilibrium (LD) structure in the ECRHS, we realised that three SNP (rs174556, rs174561, rs3834458) are sufficient to cover most of the genetic information regarding common SNP(Reference Schaeffer, Gohlke, Muller, Heid, Palmer, Kompauer, Demmelmair, Illig, Koletzko and Heinrich1). Therefore we decided to restrict genotyping to these three SNP in the BVS-II study.
Genomic DNA was extracted from ‘buffy coat’ using the FlexiGene DNA kit (Qiagen GmbH, Hilden, Germany). The samples were genotyped with the MassARRAY system using the iPLEX™ chemistry as suggested by the manufacturer (Sequenom, San Diego, CA, USA).
Briefly, genomic DNA was amplified by PCR using HotStarTaq DNA Polymerase (Qiagen). Genotyping assays were carried out by using 5 ng genomic DNA. PCR primers were used at 167 nm final concentrations for a PCR volume of 6 μl. The PCR conditions were 95°C for 15 min for a hot start, followed by denaturing at 95°C for 30 s, annealing at 56°C for 30 s, extension at 72°C for 1 min for forty-five cycles, and finally incubation at 72°C for 10 min. After PCR a shrimp alkaline phosphatase- and primer extension reaction was carried out according to the iPLEX™ reaction protocol (Sequenom). All reactions (PCR amplification, base extension) were carried out in a Tetrad PCR thermal cycler (Bio-Rad, Hercules, CA, USA). The final base extension products were treated with SpectroCLEAN resin (Sequenom) to remove salts in the reaction buffer. This step was carried out with a Multimek 96-channel autopipette (Beckman Coulter, Inc., Fullerton, CA, USA), and 16 μl resin–water suspension was added into each base extension reaction, making the total volume 26 μl. After a quick centrifugation (2000 rpm, 3 min) in an Eppendorf centrifuge 5810, 30 nl of reaction solution was dispensed onto a 384 format SpectroCHIP (Sequenom) prespotted with a matrix of 3-hydroxypicolinic acid by using a SpectroPoint nanodispenser (Sequenom). A modified Bruker Autoflex matrix-assisted laser desorption ionisation-time-of-flight mass spectrometer (Sequenom) was used for data acquisitions from the SpectroCHIP. The resulting mass spectra were analysed automatically for peak identification using the SpectroTYPER RT 3.4 software (Sequenom). For quality reasons 10 % of the spectra were checked by two independent trained individuals. No differences in spectra calling could be detected.
Statistical analysis
Allele frequencies, Hardy–Weinberg equilibrium and tests on LD for the BVS-II study data were performed with procedures ‘proc allele’ and ‘proc haplotype’ of the statistical software module SAS/Genetics of SAS version 9.1.3 (SAS Institute, Inc., Cary, NC, USA) and with the special-purpose software JLIN(Reference Carter, McCaskie and Palmer11). Hardy–Weinberg equilibriums were tested by Fisher's exact test and LD by the likelihood ratio test of allelic association and pairwise LD measures Lewontin's D′ and pairwise squared correlation r 2. The software JLIN was used to calculate pairwise LD measures D′ and r 2 and to show LD blocks graphically.
Both single SNP and haplotype regression analyses for the nine PUFA were conducted applying an additive model. Haplotype reconstruction was conducted with the procedure haplo.glm of the module haplo.stats of the R-software (R Foundation for Statistical Computing, Vienna, Austria; http://www.r-project.org). Haplo.glm is based on a special expectation-maximisation (EM)-based algorithm for estimation of haplotypes described by Schaid et al. (Reference Schaid, Rowland, Tines, Jacobson and Poland12) and was also used to estimate the associations of haplotypes on fatty acids by linear regression. Haplotypes below 1 % of haplotype frequency were combined in a rare-haplotype category. The reference category of all association analyses was the all-major three-loci haplotype, which is composed of the more frequent allele in all three loci. Normal distribution of PUFA in plasma phospholipids and in erythrocyte membranes was tested using Kolmogorov–Smirnov tests and evaluated by Q-Q plots and stem-and-leaf plots (procedure ‘proc univariate’ of SAS/STAT 9.1.3; SAS Institute, Inc., Cary, NC, USA). Severely right-skewed outcomes such as γ-linolenic acid (18 : 3n-6) and EPA (20 : 5n-3) were log-transformed. To keep the nominal level of type 1 error to 5 % despite numerous tests of associations, P values of the primary analyses were conservatively corrected by multiplying the uncorrected P value by the number of analysed outcomes (fatty acids) times the effective number of loci (SNP) according to Nyholt's method(Reference Nyholt13). Effective loci were calculated using the spectral decomposition method software SNPSpD (Queensland Institute of Medical Research, Herston, QLD, Australia; http://genepi.qimr.edu.au/general/daleN/SNPSpD/).
Results
Single nucleotide polymorphism associations with fatty acids
As a first step – before conducting haplotype analyses – we performed single SNP analyses with each PUFA derived from erythrocytes and from plasma phospholipids in the 535 and 163 participants of the BVS-II study respectively. From Table 1, it can be seen that for PUFA from erythrocytes each of the three SNP (rs174556, rs174561 and rs3834456) is highly significantly associated with dihomo-γ-linolenic and arachidonic acid, even after correction for multiple testing. Associations for adrenic acid are significant for rs174556 and rs174561, but not for rs3834458, when correction of multiple testing is accounted for. The associations of the SNP with EPA and n-3 docosapentaenoic acid were only significant before correction for multiple testing. Similar results as found for PUFA in erythrocytes were observed for PUFA in plasma phospholipids in the BVS-II study (data not shown).
Table 1 Association of single single nucleotide polymorphisms (SNP) of the fatty acid desaturase 1 fatty acid desaturase 2 (FADS1 FADS2) gene cluster with PUFA in erythrocyte membranes in the Bavarian Nutrition Survey II (n 535)

* SNP is coded additive as 0, 1, 2. Allele frequencies (in %) for the three SNP are: rs174556, C/T = 70·5/29·5; rs174561, T/C = 70·4/29·6; rs3834458, T/deletion = 67·5/32·5.
† The P value is corrected for multiple testing by multiplying the uncorrected P value by 9 (for the nine outcomes). Note that P values exceeding 1·0 after correction for multiple testing have been marked down to 1·0.
‡ Regression coefficient β from linear regression of the fatty acid (outcome) on the respective single SNP.
Estimated haplotypes
Prerequisites of valid haplotype analyses were satisfied. All three SNP (rs174556, rs174561, rs3834458) of the BVS-II study used for haplotype construction were in Hardy–Weinberg equilibrium. Minimal P value for the exact test of violation of Hardy–Weinberg equilibrium was 0·38 for rs3834458. All three SNP showed very strong LD as shown diagrammatically by pairwise Lewontin's D′ and pairwise squared correlation r 2 values in Fig. 1. Strong LD over a block of these and further SNP has also been shown previously in the ECRHS(Reference Schaeffer, Gohlke, Muller, Heid, Palmer, Kompauer, Demmelmair, Illig, Koletzko and Heinrich1).

Fig. 1 Pairwise linkage disequilibrium D′ and r 2 plots of the three common single nucleotide polymorphisms (SNP) in European Community Respiratory Health Survey and Bavarian Nutrition Survey II study participants with measures on PUFA in erythrocyte membranes (n 535). Position in bp was derived from dbSNP Build 126, based on National Center for Biotechnology Information (NCBI) Human Genome Build 36.1 (7 December 2006) of chromosome 11. Position in bp for rs174556 is 61337211, for rs174561, 61339284 and for rs3834458, 61351497.
Haplotype construction for the participants in the BVS-II study with PUFA in plasma phospholipids as well as with erythrocyte membranes resulted in three haplotypes with a haplotype frequency proportion above 1 % (see Table 2). Haplotypes were constructed from participants with no missing data on the three selected SNP and data available on the fatty acid composition (phospholipids or erythrocytes respectively). The three-loci haplotypes reflected the five-loci haplotypes (rs174544, rs174553, rs174556, rs174561, rs3834458) as estimated in the data of the ECRHS in Erfurt very well. An all-major allele haplotype (MaA, with alleles C-T-T), an all-minor allele haplotype (MiA, with alleles T-C-del), and a haplotype of major alleles except for the last locus (rs3834458; haplo.1, with alleles C-T-del) were estimated from the three SNP (rs174556, rs174561, rs3834458). In addition, the haplotype frequency proportions were about the same in the BVS-II and the ECRHS. It should be noted that the estimation of haplotypes from the available three of the five SNP resulted in a haplotype pattern at the common loci identical to that estimated in the ECRHS.
Table 2 Comparison of the five-loci haplotypes in the European Community Respiratory Health Survey (ECRHS) (n 727) and the three-loci haplotypes in the Bavarian Nutrition Survey II (BVS-II) constructed from common single nucleotide polymorphisms (SNP)
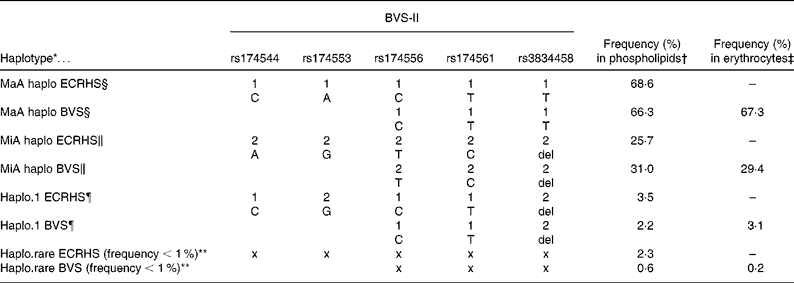
* Haplotypes reconstructed from SNP from all five SNP (ECRHS) or from three SNP (BVS-II study).
† Frequency (%) in phospholipids = % haplotype frequency in BVS-II study sample of plasma phospholipids (n 163) or serum phospholipids in ECRHS study sample (n 727).
‡ Frequency (%) in erythrocytes = % haplotype frequency in BVS-II study sample of erythrocyte membranes (n 535).
§ MaA = all-major alleles haplotype carrying only common alleles (1-1-1-1-1 or 1-1-1).
∥ MiA = all-minor alleles haplotype carrying only rare alleles (2-2-2-2-2 or 2-2-2).
¶ Haplo.1 = haplotype carrying mixture of major and minor alleles (1-2-1-1-2 or 1-1-2).
** Haplo.rare = pooled haplotype of all estimated haplotypes with a haplotype frequency below 1 %. Note that x x x x x and x x x indicate that no explicit alleles are depicted as the rare haplotype is a haplotype pooled from several haplotypes.
Fatty acid composition of plasma phospholipids and erythrocyte membranes and the FADS1 FADS2 gene cluster
Table 3 shows the mean PUFA content in erythrocytes and phospholipids in carriers of two copies of the MiA haplotype and those with two copies of the MaA haplotype. In carriers of the MiA haplotype, significantly elevated proportions of dihomo-γ-linolenic acid (20 : 3n-6) and significantly decreased proportions of arachidonic acid (20 : 4n-6) were observed in both PUFA in plasma phospholipids and in erythrocyte membranes; the results remained statistically significant after correcting for multiple testing. A significantly decreased level of adrenic acid (22 : 4n-6) in carriers of the MiA haplotype was estimated using PUFA data from erythrocytes but this was not found in plasma phospholipids.
Table 3 Mean levels of PUFA content (%) in erythrocyte membranes and plasma phospholipids for carriers and non-carriers of the all-minor alleles haplotype in the Bavarian Nutrition Survey II (BVS)

* MiA/MiA = mean level of PUFA for individuals with two copies of the all-minor alleles haplotype MiA (2-2-2/2-2-2 = T-C-del/T-C-del).
† MaA/MaA = mean level of PUFA for individuals with two copies of the all-major alleles haplotype MaA (1-1-1/1-1-1 = C-T-T/C-T-T).
‡ P values test for the difference in PUFA levels between carriers and non-carriers of the all-minor alleles haplotype. These are based on the special multiple regression model described in the Materials and methods section. This model assesses the influence of the estimated haplotypes (reported in Table 2) on the respective outcome (i.e. PUFA). P values are corrected for multiple testing according to Nyholt's procedure(Reference Nyholt13), i.e. P values are multiplied by 11 for both BVS samples (11 approximately equal to 1·191 effective loci × 9 outcomes for the BVS erythrocyte sample; 11 approximately equal to 1·163 effective loci × 9 outcomes for the BVS phospholipid sample). Note that P values exceeding 1·0 after correction for multiple testing have been marked down to 1·0.
For all remaining n-6 and n-3 fatty acids no significant differences between carriers of the MiA haplotype or any other reconstructed haplotype and carriers of two copies of the MaA haplotype could be established after correcting for multiple testing. These null findings were consistent for plasma phospholipids and erythrocytes. In the previously published analyses of PUFA in plasma phospholipids in the ECRHS (n 727) statistically significant effects of the all-minor haplotype (MiA) were shown for all nine PUFA from phospholipids except for DHA(Reference Schaeffer, Gohlke, Muller, Heid, Palmer, Kompauer, Demmelmair, Illig, Koletzko and Heinrich1). The respective analyses in the BVS-II study, however, with a sample size of n 163 resulted – regardless of the level of significance – in similar regression coefficients as in the ECRHS both in size and direction (regression coefficients not shown). Note further that this is also true for the association between the MiA haplotype on PUFA in erythrocytes (regression coefficients not shown).
We repeated the regression analyses of the effect of haplotypes on PUFA in both plasma phospholipids and erythrocytes and adjusted for total energy intake (kJ/d), physical activity (categorised as amount (h) of physical exercise per week, zero or less than 1 h per week, 1– < 2 h per week, 2– < 4 h per week, 4– < 6 h per week, 6– < 8 h per week, 8– < 10 h per week and 10+h per week) and BMI (continuous), both in separate analyses and simultaneously. We did not find any substantial differences between crude and adjusted analyses in P values, or in regression estimates (data not shown). In the 163 BVS-II participants with PUFA data on both plasma phospholipids and erythrocyte membranes, correlation coefficients for PUFA ranged from 0·05 to 0·47; for α-linolenic acid, dihomo-γ-linolenic acid and arachidonic acid correlation coefficients were 0·47 (P < 3·6 × 10− 10), 0·27 (P < 3·9 × 10− 4) and 0·05 (P < 0·51), respectively.
Discussion
This is the first replication of the previously found associations of the genetic variants in the FADS1 FADS2 gene cluster with the PUFA composition of serum phospholipids(Reference Schaeffer, Gohlke, Muller, Heid, Palmer, Kompauer, Demmelmair, Illig, Koletzko and Heinrich1) in an independent population-based study. Moreover, this is the first study showing similar differences in PUFA composition in erythrocyte membranes by polymorphisms of the FADS1 FADS2 gene cluster. In particular, we found that the MiA haplotype is negatively associated with the arachidonic acid content in plasma phospholipids as well as in erythrocyte membranes. Additionally, distinct associations between FADS1 FADS2 haplotypes and the dihomo-γ-linolenic acid (20 : 3n-6) and adrenic acid (22 : 4n-6) content in erythrocyte membranes were observed. Erythrocyte membranes are used in the present study as an easily obtainable model membrane since the biological effects of PUFA via eicosanoid formation are mediated by other cell types, including lymphocytes, platelets, or macrophages. Erythrocytes are lacking desaturase activity; however, also leucocytes show only very limited desaturase activity, if present at all.
From a physiological point of view, there are distinct differences between PUFA in plasma phospholipids and in erythrocyte membranes. The PUFA composition of plasma phospholipids is often considered as a biomarker of dietary PUFA intake, although its composition is already affected by metabolic steps, such as absorption, lipoprotein metabolism in the intestine, liver and blood, and uptake in tissue cells(Reference Arab14, Reference Kinsella, Lokesh, Broughton and Whelan15). Hepatic PUFA metabolism has a key role in the (re-) distribution of dietary PUFA to circulation via VLDL formation(Reference Kinsella, Lokesh, Broughton and Whelan15). Dietary PUFA are metabolised as energy sources (β-oxidation)(Reference Arterburn, Hall and Oken16, Reference Burdge17) and some are incorporated into structural membrane lipids. In addition, PUFA may be further desaturated and elongated by means of the desaturase–elongase enzyme system located in the endoplasmic reticulum, with the key rate-limiting enzyme Δ-6-desaturase. The highest expression rates of these enzymes are found in the liver(Reference Cho, Nakamura and Clarke18, Reference Cho, Nakamura and Clarke19). Due to the observed effects of the genetic variation in FADS1 and FADS2 genes on the composition of PUFA in plasma phospholipids a major contribution of the liver desaturase–elongase enzyme system seems likely. The activity of Δ-6 desaturase – and probably also Δ-5 desaturase – is sensitive to nutritional, metabolic and hormonal regulation(Reference Burdge17, Reference Spielmann, Bracco, Traitler, Crozier, Holman, Ward and Cotter20). Enzymes of the endogenous chain elongation and desaturation system show a higher affinity to n-3 PUFA (α-linolenic acid) as compared with n-6 PUFA (linoleic acid); however, due to a much higher proportion of n-6 PUFA (linoleic acid) in the diet, distinctly more arachidonic acid than EPA is formed(Reference Arterburn, Hall and Oken16, Reference Burdge17, Reference Spielmann, Bracco, Traitler, Crozier, Holman, Ward and Cotter20, Reference Rubin and Laposata21). The here-reported associations between desaturase haplotypes and membrane PUFA, which are highly significant for arachidonic acid and much less distinct for n-3 PUFA, are in line with the described differences in enzyme activities. The MiA haplotype of the FADS1 FADS2 gene cluster is associated with a higher proportion of the substrates and a lower proportion of the products (linoleic acid → γ-linolenic acid, dihomo-γ-linolenic acid → arachidonic acid) of these enzymes in plasma phospholipids and erythrocyte membranes. Similar effects (with respect to the expected direction, i.e. positive β-coefficient for substrates, negative β-coefficient for products) on n-3 PUFA were noted but were less pronounced and did not reach statistical significance. Although functional data are still missing, our data may indicate a lower activity of hepatic desaturases – especially Δ-5 desaturase (FADS1) catalysing the reaction of dihomo-γ-linolenic acid to arachidonic acid – in carriers of the MiA haplotype v. MaA haplotype.
Arachidonic acid and EPA, incorporated in phospholipids of cell membranes, are substrates for enzymes of the arachidonic acid pathway after their release by phospholipases(Reference Spielmann, Bracco, Traitler, Crozier, Holman, Ward and Cotter20, Reference Calder22). It has been well documented that eicosanoids derived from arachidonic acid (n-6 PUFA) are potent pro-inflammatory mediators while the effects of eicosanoids from EPA (n-3 PUFA) or dihomo-γ-linolenic acid are much less pronounced or even non-inflammatory(Reference Spielmann, Bracco, Traitler, Crozier, Holman, Ward and Cotter20, Reference Calder22). Thus, a lower proportion of arachidonic acid in cellular membranes as found for the carriers of the MiA haplotype could be interpreted as a metabolic advantage in terms of less pronounced inflammatory reactions, if induced by different stimuli. However, biological activities other than modulation of inflammation are described for PUFA as well as eicosanoids (and docosanoids) which could also influence disease risk(Reference Calder23–Reference Adam, Beringer, Kless, Lemmen, Adam, Wiseman, Adam, Klimmek and Forth28).
Concerning limitations and strengths of the study, reconstruction of the five-loci haplotypes as identified in the ECRHS study with only three SNP in the BVS-II study worked well since these SNP are in very strong LD, and an identical haplotype structure was found in the BVS-II sample. Both studies are population-based studies but conducted in different geographic regions in Germany. The genotyping success rates were high in both the BVS-II study and the ECRHS at the three SNP, rs174556 (98·9 and 94·4 %), rs174561 (97·8 and 96·4 %) and rs3834458 (98·8 and 99·2 %). Due to the lower sample size of the BVS-II study (n 535 and n 163) in comparison with the ECRHS (n 727), we cannot rule out that non-significant differences in results regarding the FADS1 FADS2 gene cluster and PUFA in plasma phospholipids are due to low power. However, despite lower power, results for dihomo-γ-linolenic and arachidonic acid are similar between ECRHS and BVS-II data.
In conclusion, in the present study we could not only replicate associations between haplotypes of the FADS1 FADS2 gene cluster and PUFA in plasma phospholipids but found also evidence for such associations in erythrocyte membranes. Especially, the association of the all-minor alleles haplotype (MiA) with a lower arachidonic acid content in cellular membrane lipids supports the hypothesis of a link between the FADS1 FADS2 gene cluster and several chronic diseases, where products of the arachidonic acid pathway are suspected to be causally involved. However, experimental data confirming the functional relevance of these genetic variants are urgently needed.
Acknowledgements
The BVS-II study was supported by funds of the Kurt-Eberhard-Bode-Stiftung and the Bavarian Ministry of Environment, Health and Consumer Protection. The present study was partly funded by the German Ministry of Education and Research (BMBF)/National Genome Research Network (NGFN) research net. Financial support by the Munich Centre of Health Sciences, which contributed to this research, is greatly acknowledged.
None of the authors has any conflict of interest.
P. R. drafted the manuscript, performed statistical analyses and was involved in interpretation. J. H., J. L. and T. I. supervised this paper and actively contributed to writing of the paper and interpretation of the results. J. L. and G. W. are principal investigators of the BVS-II and conducted together with S. H. the laboratory measurement of fatty acids in biological specimens. T. I. and N. K. did the genotyping and wrote the section on genotyping. All authors contributed to the interpretation and discussion of the results and saw and approved the final version of this manuscript.






