Cardiovascular disease accounts for approximately 17 million deaths a year, nearly one-third of the all-cause mortality. Of note, complications of hypertension accounted for 9.4 million deaths worldwide every year (World Health Organization, 2013). The common measurements of hypertension are SBP and DBP (DBP; Pickering et al., Reference Pickering, Hall, Appel, Falkner, Graves, Hill and Roccella2005). HR and PP (PP; i.e., the difference between SBP and DBP) serve as an additional predictors for cardiovascular diseases (Cooney et al., Reference Cooney, Vartiainen, Laakitainen, Juolevi, Dudina and Graham2010; Franklin et al., Reference Franklin, Khan, Wong, Larson and Levy1999). In order to better control and prevent related diseases, understanding the etiology behind the blood pressure and HR is critical. Evidence from twin studies and family-based studies suggested that genetic variance might contribute to SBP, DBP, HR, and PP variance (Hottenga et al., Reference Hottenga, Whitfield, de Geus, Boomsma and Martin2006; Rice et al., Reference Rice, Vogler, Perusse, Bouchard and Rao1989). Traditional twin study is by far the most common approach to calculate the genetic contribution for certain phenotypes by comparing intra-pair concordance, which relies on strong assumptions about the relative environmental similarity of identical (monozygotic, MZ) and fraternal (dizygotic, DZ) twins (Conley et al., Reference Conley, Rauscher, Dawes, Magnusson and Siegal2013).
Heritability, as a measure of balance between genetic and environmental contributions, would naturally vary with the environmental exposures and genetic lineage and does not have a consistent value (Elks et al., Reference Elks, den Hoed, Zhao, Sharp, Wareham, Loos and Ong2012). Earlier twin studies reported that SBP heritability ranged from 0.28 to 0.60 (Rice et al., Reference Rice, Vogler, Perusse, Bouchard and Rao1989; Slattery et al., Reference Slattery, Bishop, French, Hunt, Meikle and Williams1988; Wang et al., Reference Wang, Ouyang, Wang and Tang1990), DBP ranged from 0.32 to 0.66 (Rice et al., Reference Rice, Vogler, Perusse, Bouchard and Rao1989; Slattery et al., Reference Slattery, Bishop, French, Hunt, Meikle and Williams1988; Wang et al., Reference Wang, Ouyang, Wang and Tang1990), PP from 0.30 to 0.54 (Snieder et al., Reference Snieder, Harshfield and Treiber2003; Tarnoki et al., Reference Tarnoki, Tarnoki, Stazi, Medda, Cotichini, Nisticò and Schillaci2012) and HR from 0.50 to 0.69 (Fagard et al., Reference Fagard, Loos, Beunen, Derom and Vlietinck2003; Snieder et al., Reference Snieder, Harshfield and Treiber2003). Few of those studies compared and summarized the weighted mean value across different twin registries. Evans published heritability in six Western countries’ Caucasian twins to demonstrate a remarkable similarity of blood pressure heritability in European populations (Evans et al., Reference Evans, Van Baal, McCarron, DeLange, Soerensen, De Geus and Boomsma2003). Snieder calculated heritability between European Americans and African Americans twins to identify the heterogeneity among Caucasians and Africans (Snieder et al., Reference Snieder, Harshfield and Treiber2003). Recently, Li reported heritability of 11 metabolic phenotypes in Danish and Chinese twins, including SBP and DBP, to compare heritability between Caucasians and Asians (Li et al., Reference Li, Duan, Pang, Zhang, Duan, Hjelmborg and Kyvik2013). In addition, there are some other studies looking for age and\or sex differences in twin heritability (Evans et al., Reference Evans, Van Baal, McCarron, DeLange, Soerensen, De Geus and Boomsma2003; Vinck et al., Reference Vinck, Fagard, Loos and Vlietinck2001). However, a systematic analysis for the heritability of SBP, DBP, PP, and HR that accounts for factors such as ethnicity, age and sex is lacking.
Therefore, we aimed to identify the literature that estimated the heritability of blood pressure and HR in twin studies, and to identify and quantify the effects of potential factors that contribute to the heterogeneity of the estimates by meta-regression.
Materials and Methods
Literature Search
Selection of relevant twin studies about blood pressure and HR started with a search on electronic database PubMed (http://www.ncbi.nlm.nih.gov/pubmed/) and Embase (http://www.elsevier.com/online-tools/embase) in November, 2013. It was performed using MeSH terms of ‘blood pressure’ or ‘heart rate’ combined with the terms ‘heritability’, and we limited the search to human studies reported in the English language. A supplementary search (e.g., using the term ‘genetic contribution’ rather than ‘heritability’) was performed to identify further studies that were not captured by the original search. Abstracts of all these search results (n = 641) were examined and relevant articles (n = 178) were retrieved for review (Figure 1). Inclusion criteria were twin studies reporting quantitative estimates for blood pressure (SBP, DBP, and PP) and HR heritability.
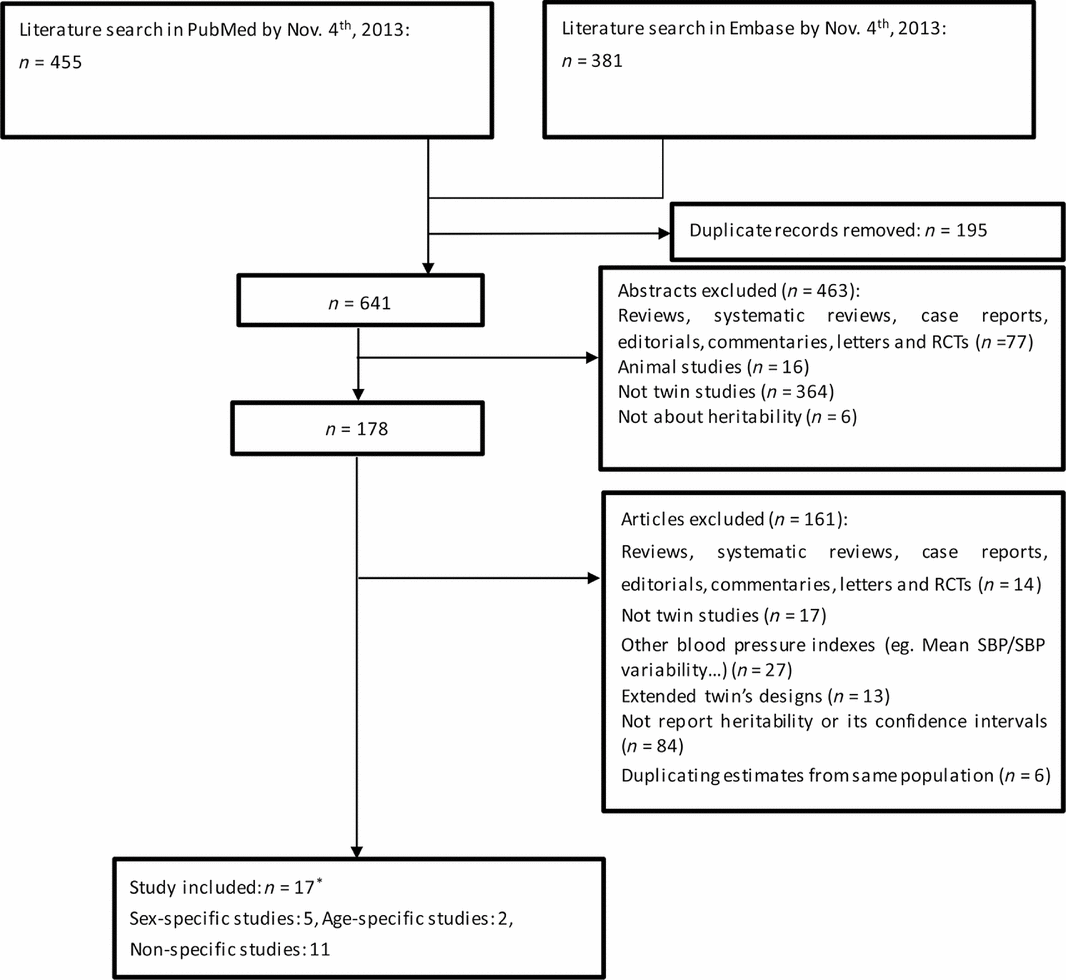
FIGURE 1 Literature search and study selection (*refers to one included study got both sex-specific results and non-specific results).
Ineligible studies were excluded from the analysis based on four exclusion criteria: (1) the estimates of 27 studies were not conventional blood pressure measurements but some other indexes such as 24-hour SBP, mean SBP or central blood pressure. (2) thirteen studies with extended twin designs (e.g., twins reared apart, twins’ blood pressure under stress or heritability measured by multiple time points) were excluded from analysis. (3) to enable a quantitative meta-analysis, measures of uncertainty for the heritability estimates were required; 84 studies without reporting heritability SE or 95% CIs were also excluded. (4) only studies from independent samples could be used for meta-analysis. Some authors used a (sub)sample of the same cohort, or reported more than one estimates under univariate and multivariate models. We selected one estimate with the most covariates included, available for largest sample, and used a variety of sensitivity analyses to address possible over adjustment or under adjustment. Finally, we had 17 studies eligible for further analysis (Figure 1).
Data Extraction
Excel 2010 was used for data extraction and quality assessment. Two trained independent researchers (Liao and Zhou) made qualitative evaluation and data extraction for each literature at the same time. Extraction data included publication information (publication year and authors), demographic characteristics (age, sex, locations and sources), study characteristics (sample size, zygosity determinants, data transformation, best fitting models and whether adjusted variables) and phenotype (SBP, DBP, PP and HR) heritability and its 95% CIs. The results were cross-referenced and any disagreements were resolved by discussion or consultation with a third evaluator (Wang).
The researchers evaluated the quality of all included studies, according to the evaluation criteria of the Agency for Healthcare Research and Quality (AHRQ; http://www.ncbi.nlm.nih.gov/books/NBK35156/). We revised the 11 items of cross-sectional prevalent studies criteria to apply in twin studies. Three assessments (items 4, 5, and 11) were removed; the remainder included items to judge the selection bias (indicate source of information, inclusion and exclusion criteria, time period used for identifying twins, explain how missing data were handled and summarize twin response rate), measurement bias (assess for quality assurance like test/retest primary outcome), and confounding bias (describe how confounding was assessed and\or controlled) (Sanderson et al., Reference Sanderson, Tatt and Higgins2007).
Statistical Analysis
The heritability of a certain phenotype was meta-analyzed when at least two independent studies were included. We calculated the SE from the reported 95% CIs of each study (Altman & Bland, Reference Altman and Bland2011), then summarized the weighted mean value of heritability and its 95% CIs by the random-effect model and whether the significant heterogeneity was based on a P value less than 0.05 (Blokland et al., Reference Blokland, de Zubicaray, McMahon and Wright2012; Borenstein et al., Reference Borenstein, Hedges, Higgins and Rothstein2011; Li et al., Reference Li, Cheng, Ma and Swan2003; Verweij et al., Reference Verweij, Zietsch, Lynskey, Medland, Neale, Martin and Vink2010). The quantity I 2 was calculated to describe the degree of heterogeneity. Subgroup analysis was performed when the studies specifically reported heritability by sex (male vs. female) or by age groups; otherwise, pooled analysis was applied to all the non-specific studies with both sex, and without any age subgroup classification.
Meta-regression was conducted to explore potential explanations such as factors regarding study designs and statistical methods for heterogeneity in SBP or DBP estimates across twin studies. Study design factors were publication year (as continuous variable), sample size (as continuous variable), ethnicity (Caucasians, African-Americans or Asians), mean age (as continuous variable) of the population and zygosity determination (DNA-based, questionnaire-based or use both DNA and questionnaire). For statistical methods, it included data transformation (binary variable: yes or no), best fitting model (AE, ACE or other methods) and adjusted covariates (binary variable: yes or no). Only one predictor was included in the regression model each time, due to the insufficient power. Funnel plots were made for assessing the publication bias. All analyses were performed with R package metafor (Viechtbauer, Reference Viechtbauer2010).
For sensitivity analyses, we examined the robustness of meta-regression results by: (1) excluding one study that might strongly affect the outcomes, because the study used Sequential Oligogenic Linkage Analysis Routine algorithm (SOLAR) instead of AE/ACE models; (2) testing whether adjusting different covariates might have any effect on meta-analysis results, because one study (Zeegers et al., Reference Zeegers, Rijsdijk, Sham, Fagard, Gielen, De Leeuw and Vlietinck2004) reported both adjusted and unadjusted model findings, and two studies (Jermendy et al., Reference Jermendy, Horváth, Littvay, Steinbach, Jermendy, Tárnoki and Osztovits2011; Wu et al., Reference Wu, Snieder, Li, Cao, Zhan, Lv and Hu2011) reported two adjusted models with different covariates.
Results
Profile of Included Studies
A total of 23 heritability estimates in SBP, 22 estimates in DBP, 7 in HR and 4 in PP were identified from 17 twin studies since year 2001 (Baird et al., Reference Baird, Osmond, MacGregor, Snieder, Hales and Phillips2001; Dalageorgou et al., Reference Dalageorgou, Ge, Jamshidi, Nolte, Riese, Savelieva and Snieder2008; De Geus et al., Reference De Geus, Kupper, Boomsma and Snieder2007; Evans et al., Reference Evans, Van Baal, McCarron, DeLange, Soerensen, De Geus and Boomsma2003; Fagard et al., Reference Fagard, Loos, Beunen, Derom and Vlietinck2003; Jermendy et al., Reference Jermendy, Horváth, Littvay, Steinbach, Jermendy, Tárnoki and Osztovits2011; Jiang et al., Reference Jiang, Jiang, Zhang, Pang, Li, Duan and Tan2012; Kennedy et al., Reference Kennedy, Rao, Botiglieri, Sharma, Lillie, Ziegler and O’Connor2005; Li et al., Reference Li, Duan, Pang, Zhang, Duan, Hjelmborg and Kyvik2013; Mutikainen et al., Reference Mutikainen, Ortega-Alonso, Alén, Kaprio, Karjalainen, Rantanen and Kujala2009; Peeters et al., Reference Peeters, Thomis, Loos, Derom, Fagard, Vlietinck and Beunen2008; Snieder et al., Reference Snieder, Harshfield and Treiber2003; Tarnoki et al., Reference Tarnoki, Tarnoki, Stazi, Medda, Cotichini, Nisticò and Schillaci2012; Vinck et al., Reference Vinck, Fagard, Loos and Vlietinck2001; Wu et al., Reference Wu, Snieder, Li, Cao, Zhan, Lv and Hu2011; Zeegers et al., Reference Zeegers, Rijsdijk, Sham, Fagard, Gielen, De Leeuw and Vlietinck2004; Zhang et al., Reference Zhang, Liu, Yu, Hong, Christoffel, Wang and Wang2009). There were four estimates in SBP, four in DBP, and two in HR reported by sex specifically; five estimates in SBP, five in DBP, and two estimates in HR reported by age subgroup separately. The rest of the estimates were non-specific studies containing both male and female data without any age subgroup classification (Table 1).
TABLE 1 Details of the Studies Reporting Blood Pressure and Heart Rate Heritability from Twin Studies*

*NA = not available; # BOTH indicated applying the DNA-based zygosity determination and questionnaire zygosity determination at the same time; †SBP = systolic blood pressure; DBP = diastolic blood pressures, HR = heart rate, PP = pulse pressure. ‡SOLAR algorithm: sequential oligogenic linkage analysis routines algorithm.
§M: male, F: female, M/F: male and female data combined.
aAdjusted covariates were age and sex.
bAdjusted covariate was sex.
cAdjusted covariate was age.
dAdjusted covariates were age, sex, ethnicity and their interaction.
eAdjusted covariates were age, sex, cholesterol ratio and body mass index (BMI).
fAdjusted covariates were age, sex, waist circumference and BMI.
gAdjusted covariates were age, sex, study site and BMI.
hAdjusted covariates were age, sex and BMI.
iAdjusted covariates were age, sex and study country.
In sex-specific studies, the sample size varied from 361 to 3,152 individuals and the mean age ranged from 25.4 to 68.5 years. The majority had data transformation in statistical analysis and they were also mostly conducted with female population and European Caucasians. Two studies used ACE as the best fitting model, and age and sex were common covariates adjusted in the models.
Demographic characters in the non-specific studies were varied among 10 countries (United Kingdom, United States, Australia, Finland, Sweden, Netherlands, Belgian, Hungary, Denmark, and China), and included Caucasians, Asians and African Americans with a mean age ranging from 14.6 to 58.6 years. Two studies stated zygosity determination was based on questionnaire, not DNA. Approximately half the studies used the ACE versus AE model as the best fitting model. One study used a SOLAR algorithm in statistical analysis; 81.2% studies adjusted for covariates commonly age, sex and body mass index (BMI) in analytical models.
Methodological Quality of the Included Studies
Seven items were used to assess the quality of 17 twin studies (Figure 2). The percentages for not reporting the source of the twins (0%), not assessing confounders (11.11%), and not reporting how to handle missing data (16.67%) were low. However, the percentages of not clearly summarizing response rate and completeness of data collection (72.22%), and not explaining any twins excluded from the study (50%) were relatively high.

FIGURE 2 The evaluations of the methodological and reporting quality of the included seventeen twin studies.
Genetic Contribution to Blood Pressure and Heart Rate
Subgroup analysis of the SBP\DBP heritability by sex was based on four studies, of which only one was conducted in male population. The SBP heritability in men was 0.58 (95% CIs: 0.46–0.70). In three studies conducted among female populations, the pooled heritability of SBP was 0.55 (95% CIs: 0.45–0.64) with significant heterogeneity (I 2 = 73.44%, p = .0068). The DBP heritability in men was 0.47 (95% CIs: 0.33–0.61) and it was 0.49 (95% CIs: 0.45–0.54) in female population. Moreover, two female studies calculated the HR heritability and reported 0.55 (95% CIs: 0.44–0.65) and 0.48 (95% CIs: 0.32–0.60) respectively.
Two studies reported five estimates by different age groups. It seemed that heritability of SBP or DBP decreased by age since adolescent, but the test for heterogeneity of all heritability across age groups remained no significance (p-value was .3459 for SBP, and .308 for DBP). The overall heritability of SBP and DBP was 0.57 (95% CIs: 0.50–0.64) and 0.55 (95% CIs: 0.48–0.62).
Among non-specific studies, 14 estimates of SBP heritability were reported in 10,613 independent twins. Heritability ranged from 0.28 to 0.71, with a weighted mean value of 0.54 (95% CIs: 0.48–0.60). Between studies heterogeneity was substantial (I2 = 82.82%, p < .001; Figure 3a). Similarly, heterogeneity was significant in DBP estimates (I2 = 78.77%, p < .001; Figure 3b) as heritability varied from 0.18 to 0.64 among 10,217 twins; the pooled DBP heritability was 0.49 (95% CIs: 0.42–0.56). Comparatively, studies about HR or PP heritability were limited. The pooled heritability of HR was 0.61 (95% CIs: 0.51–0.70); PP was 0.50 (95% CIs: 0.44–0.55).

FIGURE 3 Heritability estimates of blood pressure and heart rate in non-specific twin studies (SBP: systolic blood pressure, DBP: diastolic blood pressure, HR: heart rate, PP: pulse pressure).
Factors Associated with Blood Pressure Heritability Estimates
Zygosity determinants reached 0.05 significant levels, suggesting a significant association with SBP heritability variance. Besides, no other study design factors (publication year, mean age, ethnicity and sample size) were significantly associated with heritability variance in SBP or DBP estimates (Table 2). Meta-regression results showed that, on average, SBP heritability in DNA-based zygosity determination was 0.2047 higher (p = .0057) than in determination combining both DNA and questionnaire methods, however, the association was diminished (p = .1824) when comparing questionnaire-based versus both methods (Table 2). Another finding was that the meta-regression that found no effect of mean age on the heritability estimate confirmed the earlier results of age-specific subgroup analysis in terms of no heterogeneity across different age groups.
TABLE 2 Meta-Regression of Univariate Analyses to Identify Factors Associated with Reported SBP and DBP Heritability Estimates in Non-Specific Twin Studies

SBP = systolic blood pressure; DBP = diastolic blood pressures; SOLAR algorithm = sequential oligogenic linkage analysis routines algorithm; Bold type represents p < .05; * Indicates applying both the DNA-based zygosity determination and questionnaire zygosity determination at the same time; #All the studies in the DBP estimates were adjusted at least one covariate.
For the statistical factors, best fitting model was one explanation for SBP heritability variance. The AE model tended to have a higher estimate than the ACE model (coefficient = 0.0947, p = .0142), while the SOLAR algorithm estimate was lower than the ACE model (coefficient = -0.2443, p = .002, Table 2). Similar results were found among DBP heritability estimates (Table 2). Raw twin data transformation and covariates adjustment in the model might not affect the SBP\DBP heritability.
Since only one study applied the SOLAR algorithm, which might strongly affect the results, we conducted a sensitivity analysis by excluding that study. The association between zygosity determinants and SBP heritability was no longer significant after excluding SOLAR algorithm study (p = .5643, Table 3). Another phenomenon found in the sensitivity analysis was that sample size might affect the DBP heritability as estimates decreased by 0.0001 per individual (p = .0006, Table 3). But fewer individuals were not nominally associated with SBP heritability in meta-regression analysis (p = .7641, Table 3). SBP heritability estimates from AE variance component models were on average 0.094 higher than those from ACE models (p = .0178) so did the DBP estimates (coefficient = 0.0613, p = .0447, Table 3).
TABLE 3 Sensitivity Analysis for Meta-Regression of Univariate Analyses to Identify Factors Associated with Reported SBP and DBP Heritability Estimates in Non-Specific Twin Studies (Excluded Kennedy Study)

SBP = systolic blood pressure; DBP = diastolic blood pressures; bold type represents p < .05. *Indicates applying both the DNA-based zygosity determination and questionnaire zygosity determination at the same time. #All the studies in the DBP estimates were adjusted at least one covariate.
We conducted another sensitivity analysis testing different adjusted covariates that might affect the meta-regression results. We substituted the initial selected model with the other covariates model and found that best fitting model was one explanation for both SBP (AE vs. ACE model: coefficient = 0.0992, p = .0137; SOLAR vs. ACE model: coefficient = -0.2510, p = .0022) and DBP heritability variance (coefficient = 0.0777, p = .0335; coefficient = -0.3220, p < .0001). There were no contradictory results to the initial models with the most covariates included.
Discussion
In this meta-analysis of published twin studies, we found the pooled heritability of SBP, DBP, HR, and PP were 0.54, 0.49, 0.61, and 0.50 respectively. In addition, we have identified and quantified the possible effects of one main potential factor: the choice of final variance component model or other statistical methods, which might explain the heterogeneity in twin studies.
From the results of methodological quality assessment, the low percentage of response rate description and the rationale for exclusion might indicate a probable selection bias in most studies. This would affect the heritability estimates. Selection bias in the original study might influence the correlation coefficient estimates of MZ and DZ, thus generating biased estimates of heritability (Martin & Wilson, Reference Martin and Wilson1982). Another issue in the current meta-analysis that merits discussion was publication bias for which results were subject to experimenters’ preference, a sponsor's interests, or community expectations. Publication bias test was conducted among the literature of non-specific studies reporting of SBP and DBP heritability estimates (Figure 4); however, the number of HR and PP estimates was too small to accurately evaluate this. Test for funnel plot asymmetry of SBP estimates was -2.8768 (p = .0139). Since asymmetry might be caused by one outlier study (Kennedy et al., Reference Kennedy, Rao, Botiglieri, Sharma, Lillie, Ziegler and O’Connor2005), we excluded that study to test the funnel plot asymmetry. Publication bias remained significant (estimate = -2.8969, p = .0145) for SBP estimates. However, the p values of DBP heritability were 0.3910 and 0.5915, suggesting a low probability of bias (Viechtbauer, Reference Viechtbauer2010). Ioannidis offered several suggestions to remedy the publication bias, including use of large-scale studies, enhanced research standards and precaution to test chance of finding the true results (Ioannidis, Reference Ioannidis2005). Since there might be a publication bias in SBP estimates, large sample studies with standard methods (such as recruitment of twins, phenotypes measurement, data cleaning and analytical models) needs to be conducted.
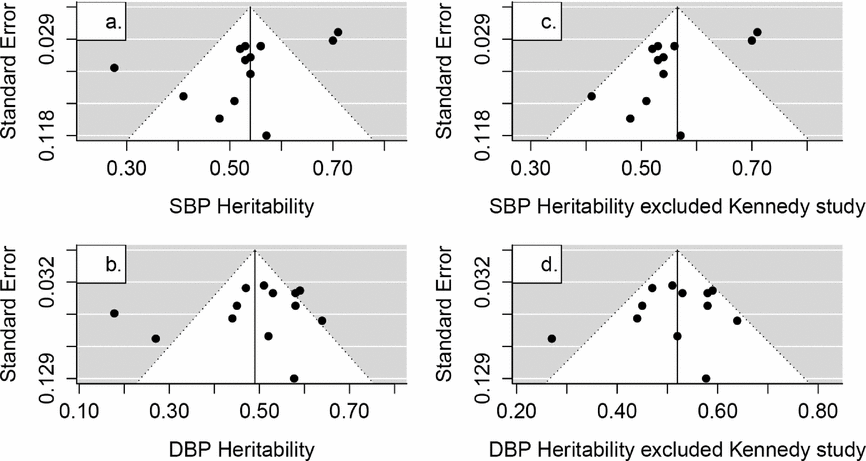
FIGURE 4 Funnel plots of the reported SBP and DBP heritability estimates in non-specific twin studies (SBP: systolic blood pressure, DBP: diastolic blood pressure).
The pooled heritability of SBP (54%) was a little more than DBP estimates (49%) in twin studies, whereas their 95% CIs overlapped, showing the 5% differences were not significant. The genetic basis contributing to heritability was found in genome-wide association studies (GWAS). A recent study found 29 independent genetic variants influence blood pressure, and collectively explained 2.2% of the phenotype variance for SBP and DBP (Ehret et al., Reference Ehret, Munroe, Rice, Bochud, Johnson, Chasman and Johnson2011). In another meta-analysis (N > 29,000) of multiple population cohorts, the most significant loci (ATP2B1) explained 0.11% SBP variance but 0.09% DBP variance (Munroe et al., Reference Munroe, Barnes and Caulfield2013; Wang & Snieder, Reference Wang and Snieder2010). Six loci for SBP and nine for DBP were discovered, of which two overlapped, yielding 13 independent genome-wide significant signals (Levy et al., Reference Levy, Ehret, Rice, Verwoert, Launer, Dehghan and van Duijn2009; Newton-Cheh et al., Reference Newton-Cheh, Johnson, Gateva, Martin, Grobbee, Onland-Moret and Bots2009). Besides, GWAS meta-analysis in East Asians found four loci and a newly discovered variant near TBX3 was related to blood pressure (Kato et al., Reference Kato, Takeuchi, Tabara, Kelly, Go, Sim and He2011). The genetic etiology behind the heritability of HR (61%) and PP (50%) was identified by GWAS: 14 new loci and 7 previously established loci had confirmed association with HR (den Hoed et al., Reference Den Hoed, Eijgelsheim, Esko, Brundel, Peal, Evans and Handsaker2013). Four new PP loci were found in related publications (Eijgelsheim et al., Reference Eijgelsheim, Newton-Cheh, Sotoodehnia, de Bakker, Müller, Morrison and O’Donnell2010; Wain et al., Reference Wain, Verwoert, O’Reilly, Shi, Johnson, Johnson and van Duijn2011). By comparing twin studies and GWAS, it was suggested that the identified loci generally accounted for a small fraction of the genetic variance estimated from twin studies. The large gap between those heritability estimates might be bridged by progress in increasing the coverage of the genome, increasing study sample size through meta-analyses and advancing statistical methods. Further analysis of gene-environment interaction could be expected to be account for the missing heritability of blood pressure (Kaprio, Reference Kaprio2012).
Association between zygosity determination and SBP heritability was no longer significant after excluding the Kennedy et al. (Reference Kennedy, Rao, Botiglieri, Sharma, Lillie, Ziegler and O’Connor2005) study. This result of the sensitivity analysis showed that the zygosity determination effect might not be a real effect, because it was explained by a small study that used different software to estimate heritability. Although another scholar thought questionnaire-based zygosity determination might underestimate heritability, because such questionnaire were upon subjective assessment, any non-differential misclassification error would inflate the E component and reduce the additive genetic component (Elks et al., Reference Elks, den Hoed, Zhao, Sharp, Wareham, Loos and Ong2012). Our study might not support the view of zygosity determination affecting blood pressure heritability variance.
We found that SOLAR, AE or ACE models were related to the SBP and DBP heritability. SOLAR is a flexible and extensive software package for genetic variance components analysis, including linkage analysis, quantitative genetic analysis, and covariate screening (Fava et al., Reference Fava, Ricci, Burri, Minuz and Melander2008; Hassan et al., Reference Hassan, Bayoumi, Lopez-Alvarenga, Snieder, Jaju, Al-Yahyaee and Albarwani2009; Sung et al., Reference Sung, Lee and Song2009). A workshop simulated a set of data to compare twin heritability by SOLAR or AE\ACE models. It was suggested that AE\ACE models would be less biased compared to SOLAR without known common environment contribution (C) or under the assumption C > 0 in twin studies. But when given C = 0, SOLAR would be less biased than AE\ACE models (Almasy & Blangero, Reference Almasy and Blangero1998; Boker et al., Reference Boker, Neale, Maes, Wilde, Spiegel, Brick and Bates2011). Not surprisingly, heritability (variance attributed to the A component) was higher in studies reporting the AE model than the ACE model, presumably because the variance that would have been attributed to C is reallocated to components A rather than E in these analyses.
Sensitivity analysis showed that sample size might be negatively associated with heritability variance in DBP but not SBP. A probable explanation was that the large Wu et al. (Reference Wu, Snieder, Li, Cao, Zhan, Lv and Hu2011) study (with a sample size of 2,076), which reported a fairly low DBP heritability of 0.27, could detect a significant effect of C given their large sample size, and therefore used an ACE rather than an AE model. As such, although it appears that the sample size had a negative effect on the DBP heritability, this may have been simply a function of model choice (AE vs. ACE model), as we have also found.
It was important to emphasize that there were several strengths and limitations in this study. The first and foremost strength was that we completed the whole comprehensive process of the systematic review and meta-regression in twin studies with wide range of the literature search, methodological evaluation and bias analysis to get more reliable results (Borenstein et al., Reference Borenstein, Hedges, Higgins and Rothstein2011). Second, we first systematically analyzed the factors that might be associated with SBP and/or DBP heritability in twins, which could be confirmed or interpreted by further genetic studies. However, there was a limitation to our study based on the published literature, that is, other unreported factors might have an impact of heritability estimates. For example, a recent meta-analysis of twins found that BMI heritability was sensitive to the study population's average BMI, gross domestic product (GDP) and GDP growth (Min et al., Reference Min, Chiu and Wang2013). Another limitation was we could only include one predictor at a time in the meta-regression due to the power consideration (Higgins et al., Reference Higgins, Thompson, Deeks and Altman2002; Schmid et al., Reference Schmid, Stark, Berlin, Landais and Lau2004).
In conclusion, in our meta-analyses that aggregated the results of a number of previous twin studies, we provided more robust estimates of the genetic influences on SBP\DBP\HR\PP. Because our analyses averaged estimates over samples of different sizes and demographic make-up, our findings were likely to be more generalizable than the individual study. The results indicated that heritability estimates based on twin studies of both SBP and DBP are around 50%. Our study confirmed that the blood pressure heritability calculation should pay attention to statistical methods which estimate genetic contribution. Reporting the AE rather than the ACE model, the variance due to C ended up in A (but not in E). So indeed, the AE model might overestimate heritability if a small contribution of C existed.
Acknowledgments
This study was supported by China Medical Board (01-746), National Natural Science Foundation of China (81202264) and the Specific Research Project of Health Public Service, Ministry of Health, China (201002007).









