Introduction
Species classification and delimitation within the fungi received a strong impetus when molecular phylogenetic methods emerged (Blackwell Reference Blackwell2011; Hibbett & Taylor Reference Hibbett and Taylor2013). Molecular methods have helped to solve several long-standing questions about higher-level taxonomic relationships among lichen-forming fungi (e.g. see James et al. Reference James, Kauff, Schoch, Matheny, Hofstetter, Cox, Celio, Gueidan, Fraker and Miadlikowska2006; Thell et al. Reference Thell, Crespo, Divakar, Kärnefelt, Leavitt, Lumbsch and Seaward2012; Miadlikowska et al. Reference Miadlikowska, Kauff, Högnabba, Oliver, Molnar, Fraker, Gaya, Hafellner, Hofstetter and Gueidan2014; Resl et al. Reference Resl, Schneider, Westberg, Printzen, Palice, Thor, Fryday, Mayrhofer and Spribille2015). However, applying molecular methods also creates new challenges and discussions are ongoing about how to crosslink phenotypic (including morphology, anatomy and secondary chemistry) information with DNA sequence data when drawing taxonomic conclusions such as the description of new lineages (Grube & Kroken Reference Grube and Kroken2000; Crespo & Lumbsch Reference Crespo and Lumbsch2010; Hibbet & Taylor 2013).
The issue of reconciling molecular and phenotypic evidence to develop taxonomic hypotheses for individual species may be simplified to three different scenarios: 1) molecular and phenotypic evidence both support taxonomic conclusions; 2) taxonomic conclusions are supported by molecular but not phenotypic characters; and 3) phenotypic evidence supports taxonomic conclusions while molecular methods do not.
Not all characters may coincide with a taxonomic hypothesis. Nevertheless, a broad consensus exists that multiple independent lines of evidence in support of taxonomic conclusions should be the desired goal of every modern taxonomic study. Lichenologists, however, are often faced with the challenge of phylogenetic and phenotypic data being inconsistent (e.g. Baloch & Grube Reference Baloch and Grube2009; Altermann et al. Reference Altermann, Leavitt, Goward, Nelsen and Lumbsch2014; Lücking et al. Reference Lücking, Dal-Forno, Sikaroodi, Gillevet, Bungartz, Moncada, Yánez-Ayabaca, Chaves, Coca and Lawrey2014; Singh et al. Reference Singh, Dal Grande, Divakar, Otte, Leavitt, Szczepanska, Crespo, Rico, Aptroot and Cáceres2015). With more sequences becoming available, species boundaries traditionally circumscribed by phenotypic characters were challenged and it was proposed that extant taxa numbers were systematically underestimated when relying on phenotypic characters alone (Crespo & Lumbsch Reference Crespo and Lumbsch2010; Lumbsch & Leavitt Reference Lumbsch and Leavitt2011). However, there are also cases where molecular methods have revealed that phenotypic differences might overestimate species diversity (Xanthoparmelia: Leavitt et al. 2011 Reference Leavitt, Johnson and St. Claira , Reference Leavitt, Johnson, Goward and St. Clairb ; Bryoria section Implexae: Myllys et al. Reference Myllys, Velmala, Holien, Halonen, Wang and Goward2011; Velmala et al. Reference Velmala, Myllys, Goward, Holien and Halonen2014).
While phenotypic differences between species can be subtle (Spribille et al. Reference Spribille, Resl, Ahti, Pérez-Ortega, Tønsberg, Mayrhofer and Lumbsch2014) they still represent a valuable source of information with which to formulate hypotheses on how to delimit species, especially in groups where phylogenetic data remain fragmentary. One such group, which has been subjected to rather few molecular phylogenetic studies, is the family Physciaceae (e.g. Lohtander et al. Reference Lohtander, Källersjö, Moberg and Tehler2000, Reference Lohtander, Myllys, Källersjö, Moberg, Stenroos and Tehler2009; Wedin et al. Reference Wedin, Döring, Nordin and Tibell2000; Grube & Arup Reference Grube and Arup2001; Helms et al. Reference Helms, Friedl and Rambold2003; Crespo et al. Reference Crespo, Blanco, Llimona and Ferencová2004; Kaschik Reference Kaschik2006; Nadyeina et al. Reference Nadyeina, Grube and Mayrhofer2010; Kondratyuk et al. 2014 Reference Kondratyuk, Lőkös, Kim, Joeng, Kondratiuk, Ou and Hura ). In this family, taxa are delimited mainly on the basis of secondary chemistry, asci and ascospores. The latter particularly have been emphasized as important and, consequently, ascospores are usually summarized into different ‘types’ (Poelt & Mayrhofer Reference Poelt and Mayrhofer1979; Mayrhofer & Moberg Reference Mayrhofer and Moberg2002; Sheard Reference Sheard2010). A thorough phylogenetic treatment of the Physciaceae including members of all known genera and sequences of multiple gene fragments to test these concepts is still missing. We focus here on two examples of closely related taxa with unclear taxonomy from the genus Rinodina with Physcia- and Physconia-type spores, and various secondary compounds. The combined analysis of phenotypic and molecular characters also allows us to provide integrated hypotheses of species taxonomy and nomenclatural status.
The first case was presented to us in the course of studying corticolous Rinodina specimens collected by GT and TT from Japan. Fertile, esorediate collections were found showing close morphological, anatomical and chemical similarities to the sorediate species Rinodina degeliana Coppins. Rinodina degeliana is scattered throughout the Northern Hemisphere. It occurs in Europe (Coppins Reference Coppins1983; Tønsberg Reference Tønsberg1992; Mayrhofer & Moberg Reference Mayrhofer and Moberg2002; Giavarini et al. Reference Giavarini, James and Purvis2009), North America (Sheard Reference Sheard2010; Lendemer et al. Reference Lendemer, Tripp and Sheard2014) and Japan (GT, TT, J. W. Sheard, pers. obs.). The esorediate specimens were subsequently found to be phenotypically identical to the holotype of Rinodina subparieta (Nyl.) Zahlbr., a species considered to be endemic to Japan.
The treatment of otherwise identical sorediate or esorediate morphs has been discussed by Tønsberg (Reference Tønsberg1992), and more recently by Brodo & Lendemer (Reference Brodo and Lendemer2012), among others. For a full literature list and exhaustive discussions, please consult the references in these two papers. Suffice it to say that there is no consensus on how different morphs of lichen taxa should be recognized. Most instructive has been the molecular study of Tehler et al. (Reference Tehler, Irestedt, Bungartz and Wedin2009), of a single species aggregate within the genus Roccella where different taxa were found to be either variable or uniform with respect to their reproductive morphs. Therefore, it appears that all apparent pairs of fertile and vegetative morphs must be studied in detail in order to decide their taxonomic status. This leads us to question if the names Rinodina degeliana and R. subparieta should both continue to be recognized as distinct species distinguished by the presence or absence of soralia.
The second example is Rinodina mniaraea (Ach.) Körb., a terricolous taxon that has been studied in detail by Timpe (Reference Timpe1990) and Ertl (Reference Ertl2000). It is found in cold and moist alpine dwarf shrub and grassland habitats on bare soil, bryophytes and plant remnants (Mayrhofer & Moberg Reference Mayrhofer and Moberg2002; Sheard Reference Sheard2010). Within R. mniaraea several infraspecific taxa have been recognized, differing in chemistry (Mayrhofer & Moberg Reference Mayrhofer and Moberg2002). All these taxa regularly form apothecia with Lecanora-type asci, possess large Physcia-type ascospores and frequently contain variolaric acid. They have been accepted as varieties in a treatment of the genus Rinodina (Mayrhofer & Moberg Reference Mayrhofer and Moberg2002), although Sheard (Reference Sheard2010) did not accept the use of varietal status. However, the names R. cinnamomea (Th. Fr.) Räsänen and R. mniaraeiza (Nyl.) Arnold already existed at the species rank for two of the varieties. How should these entities be referred to and which taxonomic rank is appropriate for them?
Here we present the results of a study using collections of R. degeliana and R. subparieta from multiple localities within their known Northern Hemisphere range and material of the three varieties of R. mniaraea from the European Alps and western North America. We performed detailed morphological, anatomical and chemical analyses of material from those species. Additionally, we reconstructed maximum likelihood and Bayesian phylogenies of numerous Rinodina taxa including R. degeliana, R. subparieta, R. mniaraea var. mniaraea, R. mniaraea var. mniaraeiza and R. mniaraea var. cinnamomea based on sequences from nuclear and mitochondrial gene fragments. We performed species delimitation methods using a Bayesian implementation of the General Mixed Yule Coalescence model (Reid & Carstens Reference Reid and Carstens2012) to find evidence for our two aims: 1) to examine species delimitation within these closely related sets of taxa in order to re-evaluate esorediate and sorediate morphs (in the case of R. degeliana/subparieta) and different chemotypes (R. mniaraea s. lat.), and 2) to decide on the status of the names R. degeliana, R. subparieta and the varieties of R. mniaraea s. lat.
Materials and Methods
SC, TS, GT, TT and James Lendemer provided fresh collections for molecular analysis. For Rinodina degeliana and R. subparieta, procedures for morphological and anatomical examination, and photography, follow those of Sheard et al. (Reference Sheard, McCune and Tønsberg2014). For R. mniaraea s. lat., we provide revised descriptions based on Timpe (Reference Timpe1990). Spore measurements for R. subparieta/R. degeliana are quoted as the 25%–75% range around the median with the 5% and 95% outliers in brackets. For R. mniaraea s. lat the average spore size and the minimum and maximum range are given. Thin-layer chromatography was carried out according to the methods of Culberson & Kristinsson (Reference Culberson and Kristinsson1970) and Culberson (Reference Culberson1972) with later modifications.
As a basis for our study we selected sequences from the dataset of Nadyeina et al. (Reference Nadyeina, Grube and Mayrhofer2010) containing a broad range of Physciaceae species with different ascospore types and secondary metabolites. We were especially interested in the relationships of R. degeliana and R. subparieta with other Rinodina species possessing Physcia-type ascospores and the secondary compound atranorin. In addition to the ten samples from R. degeliana and two from R. subparieta, we also included eight specimens of R. mniaraea from its three varieties, all possessing Physcia-type ascospores and sometimes atranorin. Furthermore, we sequenced R. freyi H. Magn. and R. efflorescens Malme with Physcia-type spores, but both lacking atranorin. To complete our dataset we included samples of the previously unsequenced or rarely sequenced species R. trevisanii (Hepp) Körb. and Phaeorrhiza nimbosa (Fr.) H. Mayrhofer & Poelt, and also the hitherto unplaced Oxnerella safavidiorum S. Y. Kondr. et al. (Table 1).
Table 1 ID, origin, species and used loci of the present molecular studies. IDs indicate laboratory-tracking numbers and also correspond to numbers used in Fig. 1 and the main text. Newly published sequences generated in this study are indicated in bold
DNA isolation, PCR amplification and sequencing
Thallus squamules or apothecia were placed in 1·5 ml Eppendorf tubes, frozen at −80°C and ground to powder in a Retsch cell grinder. We directly applied lysis buffer to the sheared cells. DNA extraction followed the QIAmp DNA Investigator Kit protocol according to the manufacturer’s instructions. We eluted the nucleic acids in 50 µl elution buffer and used them undiluted for subsequent polymerase chain reactions (PCR). For each sample we sequenced the internal transcribed spacer regions 1 and 2 with the embedded 5.8S region of the ribosomal DNA (ITS) and the mitochondrial ribosomal small subunit (mtSSU). PCR was performed with PuReTaq Ready-To-Go PCR beads using primer pairs ITS1F/ITS4 (Gardes & Bruns Reference Gardes and Bruns1993; White et al. Reference White, Bruns, Lee and Taylor1990) and mtSSU1:mtSSU3R (Zoller et al. Reference Zoller, Scheidegger and Sperisen1999) for ITS and mtSSU, respectively. PCR products were checked for the correct size on ethidium bromide-stained agarose gels and purified using the Omega E.Z.N.A Cycle Pure Kit according to the manufacturer’s instructions. Purified PCR products were sequenced by Microsynth (Vienna).
Alignment and phylogenetic reconstruction and tree selection
Raw sequences were subjected to manual quality control with BioEdit 7.2.5 (Hall Reference Hall1999) and only high quality sequences were used for phylogenetic analyses. Sequences were aligned and concatenated using our in-house script pipeline (Resl et al. Reference Resl, Schneider, Westberg, Printzen, Palice, Thor, Fryday, Mayrhofer and Spribille2015; Resl Reference Resl2015). We aligned sequences with MAFFT v.7 (Katoh & Standley Reference Katoh and Standley2013) employing the –genafpair algorithm. Alignments were checked manually with BioEdit 7.2.5 and only obvious errors (when MAFFT placed single nucleotides on either end of the alignments introducing long gaps) were corrected. These alignments were then used, without excluding any sites. Data matrices and trees are deposited at Treebase.org under study ID: 19205.
We created gene trees for the two gene fragments with RAxML v.8 (Stamatakis Reference Stamatakis2014). For each gene fragment we set RAxML to generate 500 fast bootstrap replicates. We partitioned the ITS dataset into the ITS1, 5.8S and ITS2 components and used PartitionFinder 1.1.1 (Lanfear et al. Reference Lanfear, Calcott, Ho and Guindon2012) with a greedy search strategy under a BIC criterion to determine the optimal number of partitions and substitution models for RAxML. Accordingly, the dataset was divided into a 5.8S and ITS1/ITS2 partition with GTRGAMMA respectively. For the mtSSU dataset we used one partition and the GTRGAMMA substitution model. We identified topological conflicts among gene trees with compat.py (Kauff & Lutzoni Reference Kauff and Lutzoni2002) based on a support threshold of 70. Conflict-free single locus datasets were combined in a concatenated RAxML analysis. Again we used PartitionFinder to identify the optimal number of partitions and substitution models. Accordingly we used two partitions (ITS1/ITS2/mtSSU and 5.8S) with a GTRGAMMA substitution model and generated 500 fast bootstrap replicates.
We also performed Bayesian phylogenetic reconstruction of the ITS dataset using BEAST 2.2.1 (Bouckaert et al. Reference Bouckaert, Heled, Kühnert, Vaughan, Wu, Xie, Suchard, Rambaut and Drummond2014). We determined the best site model and partitioning scheme for the ITS dataset (ITS1: GTR+G, 5.8S: TrNef+G, ITS2: GTR+G) with PartitionFinder 1.1.1 according to the BIC criterion and used a relaxed log-normal clock model for each partition. We linked the tree model and subjected it to a Yule tree prior. The MCMC chain was run for 107 generations and every 1000th tree was retained. Convergence of model parameters was assessed using Tracer 1.6 (Rambaut et al. Reference Rambaut, Suchard, Xie and Drummond2014) and considered sufficient for ESS values larger than 200. After discarding the first 20% of trees from the posterior tree sample as burn-in, we created a maximum clade credibility tree with TreeAnnotator 2.2.1 included in the BEAST package and randomly selected 100 trees using bash scripts. The maximum clade credibility tree and the set of 100 random trees were used in all further analyses.
Species delimitation using General Mixed Yule Coalescence
We performed species delimitation with the statistical programming language R (R Development Core Team 2013) using the R package bGMYC (Reid & Carstens Reference Reid and Carstens2012). This package provides a Bayesian implementation of the general mixed Yule-coalescence model (Pons et al. Reference Pons, Barraclough, Gomez-Zurita, Cardoso, Duran, Hazell, Kamoun, Sumlin and Vogler2006). Based on a given tree sample, bGMYC models the contribution of between-species divergence (Yule model component) and within-species coalescence events (Coalescent model component) (Pons et al. Reference Pons, Barraclough, Gomez-Zurita, Cardoso, Duran, Hazell, Kamoun, Sumlin and Vogler2006), which give a hypothesis for species delimitation. To account for phylogenetic uncertainty in our dataset we performed all bGMYC analyses on a set of 100 randomly selected trees from the BEAST posterior tree sample (see above). We used the function bgmyc.multiphylo() to sample 50 000 steps with a burn-in of 40 000 and the thinning parameter set to 100 for each tree. We then created a co-assignment probability heat map as implemented in the bGMYC function spec.probmat(). To determine consensus partitions from the probability heat map, we employed a k-medoid clustering approach as described in Ortiz-Álvarez et al. (Reference Ortiz-Álvarez, de los Ríos, Fernández-Mendoza, Torralba-Burrial and Pérez-Ortega2015). We used the function pamk() in the R package fpc (Hennig Reference Hennig2013) to create groups from the bGMYC co-assignment matrix. Pamk() uses a partitioning around medoid (PAM) algorithm, which randomly selects data points from the co-assignment matrix as centres and clusters other data points around those centres according to their distance. The number of clusters is determined by optimum average silhouette width (Hennig Reference Hennig2013).
Results
Phylogenetic analyses
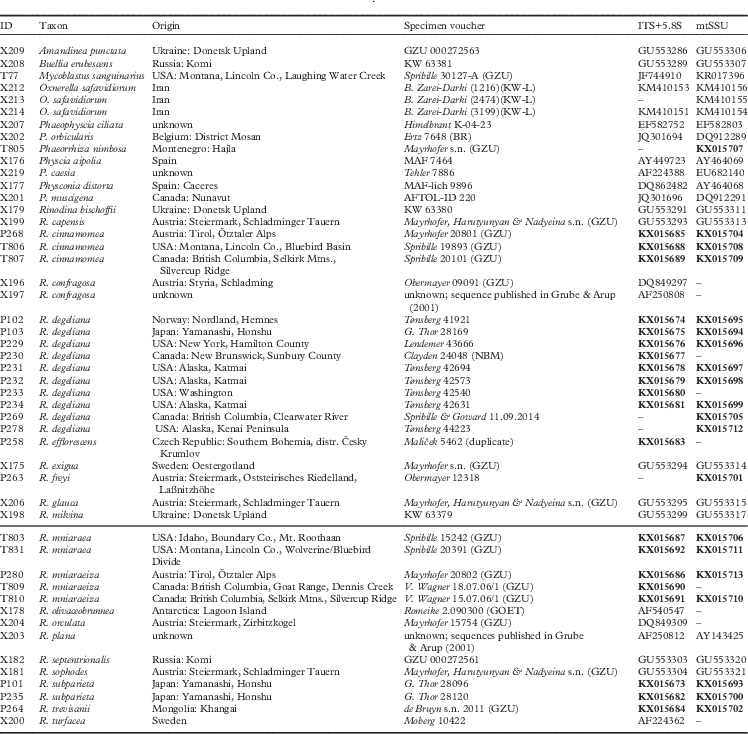
We obtained 40 sequences from 24 isolates. Together with the sequences downloaded from GenBank, our dataset consisted of 49 samples and a total of 84 sequences. All sequences used, including those newly published, are summarized in Table 1. Results from our maximum likelihood analyses are depicted in Fig. 1A–C. We identified one conflict between the ITS (Fig. 1B) and mtSSU (Fig. 1C) gene trees coming from specimen P280. As ITS is an important fungal barcode (Schoch et al. Reference Schoch, Seifert, Huhndorf, Robert, Spouge, Levesque and Chen2012) and our species delimitations were based on the ITS dataset, we excluded mtSSU sequences of these specimens from the concatenated phylogenetic analysis.
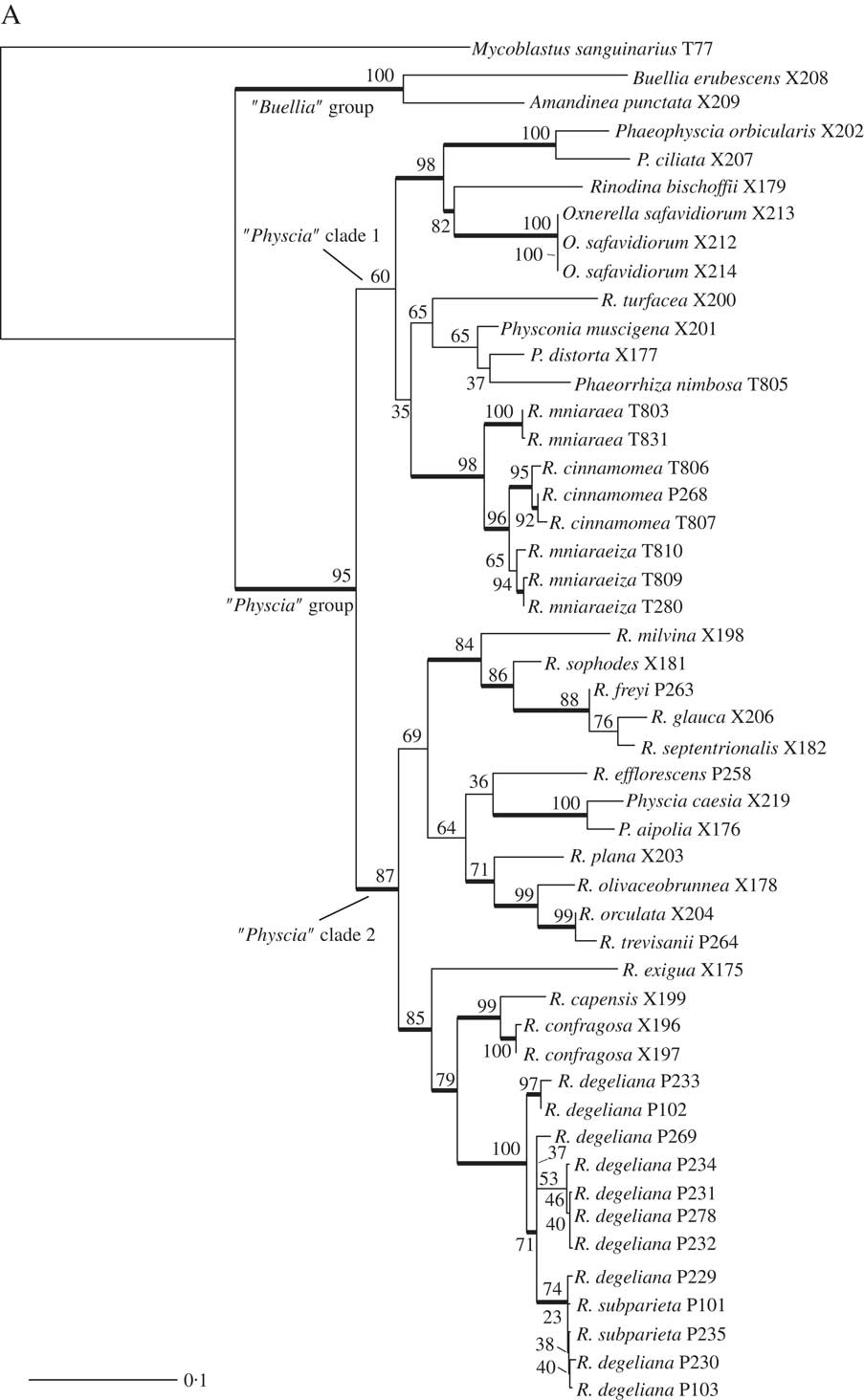
Fig. 1A A, maximum-likelihood phylogenetic hypothesis of the concatenated ITS and mtSSU dataset. Branch support is provided as bootstrap values. Branches with high support (bootstrap value ≥70) are indicated in bold.
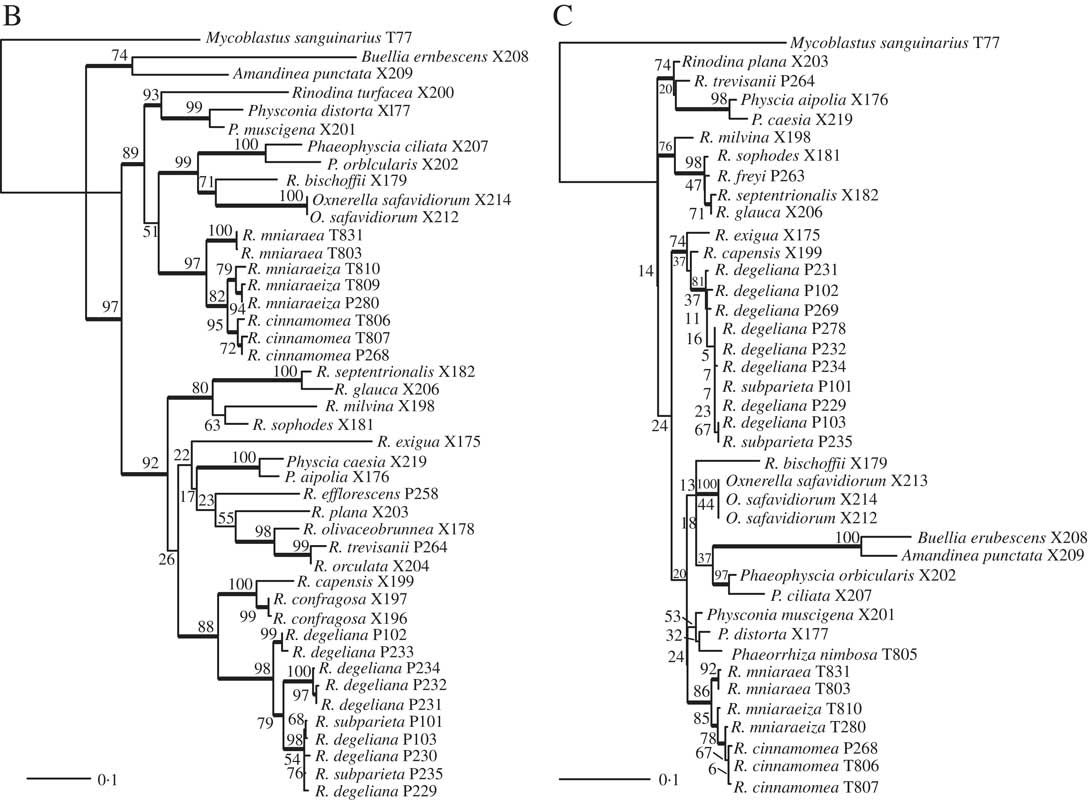
Fig. 1B and C (continued). B, maximum-likelihood ITS gene tree. Branch support is provided as bootstrap values. Branches with high support (bootstrap value =≥70) are indicated in bold. C, maximum-likelihood mtSSU gene tree. Branch support is provided as bootstrap values. Branches with high support (bootstrap value =≥70) are indicated in bold. Scales show nucleotide substitutions per site.
The deepest split in our concatenated maximum likelihood tree (Fig. 1A) corresponded to the “Buellia” and “Physcia” groups already reported by Nadyeina et al. (Reference Nadyeina, Grube and Mayrhofer2010). The “Buellia” group, here represented by Amandinea punctata (Hoffm.) Coppins & Scheid. and Buellia erubescens Arnold was well supported (100% bootstrap support (BS)). The “Physcia” group, which received high support (95% BS) was divided into two clades, hereafter referred to as “Physcia” clade 1 and 2 (Fig. 1A). “Physcia” clade 1 was only moderately supported (60% BS). It contained all samples of Rinodina mniaraea s. lat., which formed a well-supported monophyletic group (98% BS). All samples of the varieties R. mniaraea var. mniaraea and R. mniaraea var. cinnamomea, respectively, formed well-supported clades (>70% BS). The clade containing R. mniaraea var. mniaraeiza was supported with 65% BS. Another well-supported (98% BS) lower level clade of “Physcia” clade 1 consisted of Phaeophyscia ciliata (Hoffm.) Moberg, P. orbicularis (Neck.) Moberg, Rinodina bischoffii (Hepp) A. Massal. and Oxnerella safavidiorum. The latter was resolved as sister to Rinodina bischoffii with high support (82% BS). The third subclade of “Physcia” clade 1, containing Rinodina turfacea (Wahlenb.) Körb., Physconia muscigena (Ach.) Poelt, Phaeorrhiza nimbosa and Physconia distorta (With.) J. R. Laundon, received poor support (65% BS).
“Physcia” clade 2 was well supported (87% BS) and contained all samples of R. degeliana and R. subparieta. They formed a monophyletic group (100% BS) with two subclades, both with high support (71% and 97% BS). Sister to the R. subparieta/degeliana clade we recovered R. capensis Hampe and R. confragosa (Ach.) Körb., which form a clade that was preceded by the divergence of Rinodina exigua (Ach.) Gray, all also containing atranorin.
Sister to the above group, the “Physcia” clade 2 also contained a moderately supported clade (69% BS) consisting of Rinodina milvina (Wahlenb.) Th. Fr., R. glauca Ropin, R. septentrionalis Malme, R. freyi, R. sophodes (Ach.) A. Massal., R. efflorescens Malme, R. olivaceobrunnea C. W. Dodge & G. E. Baker, R. orculata Poelt & M. Steiner, R. plana H. Magn, R. trevisanii (Hepp) Körb., Physcia aipolia (Ehrh. ex Humb.) Fürnr. and P. caesia (Hoffm.) Fürnr., all lacking atranorin except for the Physcia species.
The BEAST analyses of the ITS dataset led to stable parameter values (ESS>200) according to Tracer. We only present the BEAST maximum clade credibility tree here (Fig. 2), with posterior probability node support indicated by different colours.
Fig. 2 Phylogenetic hypothesis and species delimitation results of the ITS dataset according to the Bayesian General Mixed Yule-Coalesence model. The tree displayed is the maximum clade credibility tree from the BEAST analysis. Node support is provided as coloured circles. Probabilities of the bGMYC probability map are indicative for the chance that tips could be assigned to one species. The pamk clusters refer to grouping based on k-medoids. bGMYC and pamk clustering was based on a tree sample of 100 randomly selected trees from the BEAST posterior distribution of trees.
Species delimitation analysis
Based on the Bayesian mixed Yule-coalescent analysis for species delimitation (Reid & Carstens Reference Reid and Carstens2012), we detected significant levels of genetic structure in Rinodina degeliana and R. subparieta, as well as in R. mniaraea s. lat. The pamk clustering approach (see above) detected 21 clusters in the whole dataset. Rinodina degeliana and R. subparieta samples form a total of three clusters (cluster numbers 1–3; see Fig. 2). The two specimens of R. subparieta are assigned to cluster 1, together with three R. degeliana samples.
Rinodina mniaraea s. lat. also forms three clusters (cluster numbers 6–8; Fig. 2), corresponding to the chemotypes R. mniaraea var. mniaraea, R. mniaraea var. mniaraeiza and R. mniaraea var. cinnamomea.
Morphological investigation
We undertook an a posteriori examination of ascospore size and structure, and soredium size between the R. subparieta/R. degeliana clades (Fig. 2) distinguished by the molecular analysis. This was necessarily incomplete due to the absence of ascospores in non-fertile specimens.
The esorediate Japanese collections (P101, P235) had very well-developed thalli and, perhaps for this reason, ascospores were also fully developed (Physconia-type) and slightly longer ((20·5–)22·5–24·5(–25·5)×(10·5–)11·0–11·5(–12·5) µm, n=66, l/w ratio (1·8–)2·0–2·2(–2·3)) than those found in the type of R. subparieta ((19·5–)21·0–23·5(–24·5)×(10·0–)11·0–12·0(–13·0) µm, n=40, l/w ratio (1·7–)1·8–2·0). The Physconia-type internal ascospore structure, well-developed torus, and darkly pigmented and strongly ornamented ascospore walls are very similar in all these specimens. It should also be noted that Nylander (Reference Nylander1900) measured even larger spores, up to 27 µm long, in the type. In addition, it is notable that the fully developed ascospores of the types of R. subparieta and R. degeliana are very similarly sized.
Fertile, sorediate Japanese specimens (representative in molecular study: P103) possessed ascospores that were on average smaller than those of esorediate specimens and apparently always belonged to the Physconia-type at maturity, measuring (17·0–)18·5–22·0(–23·5) × 10·0–11·0(–12·5) µm (n=55), l/w ratio (1·5–)1·7–2·0(–2·1). Immature ascospores have broad lumin canals (septal wall thickenings), some with additional apical wall thickening (Physcia-like, Fig. 3D). Young ascospores sometimes lose the apical thickening during further development and finally become constricted at the septum. Lumina are then uniformly rounded and thin walled (Milvina-like). Measurements for ascospores in specimens with incompletely developed, Physcia-type spores tended to be shorter and comparable with those given by Sheard (Reference Sheard2010), (15·0–)18·5–19·5(–23·0)×(8·5–)10·0–11·0(–13·0) µm, for North America. The single fertile eastern North American specimen analyzed (Lendemer 43666; P229, Fig. 3A) had poorly formed apothecia with infrequent ascospores that were not measured since they were either too young or too old. Most of its apothecia had abnormally well-developed proper exciples, making them appear lecideine. However, the margins were thalline below, with typically deep cortices filled with atranorin crystals.
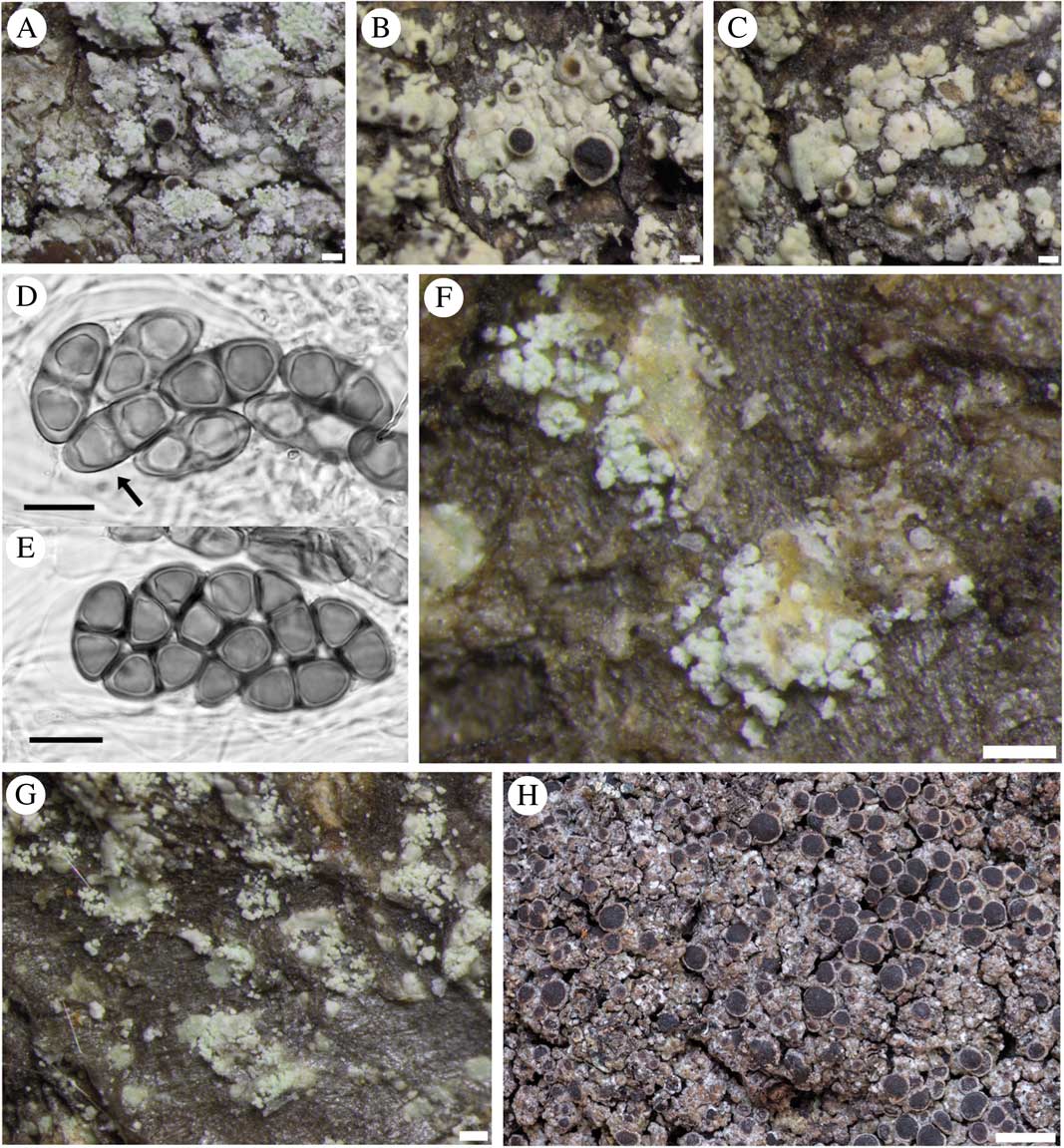
Fig. 3 Rinodina subparieta (A–G). Fertile sorediate morph (Lendemer 43666, GZU); B & C, fertile esorediate morph (Thor 28096); D, submature Physconia-type ascospores (arrow pointing to Physcia-like ascospore with narrow lumin canals and apical wall thickening) (Tønsberg 22502, BG); E, mature Physconia-type ascospores with broad lumin canals, no apical wall thickening and prominent tori at the septa in various optical sections (Tønsberg 22502, BG); F, labriform soralia arising from slightly upturned areole margins (Tønsberg 42540); G, sterile morph showing labriform soralia (Tønsberg 41921); H, R. mniaraea showing typical matt, rugose grey-brownish thallus. Scales: A–C, F & G=200 µm; D & E=10 µm; H=1 mm.
Alaskan specimens (P231, P232, P234, pamk cluster 3) were all sorediate and mostly sterile. The single fertile specimen (Tønsberg 42694; P231) analyzed from Alaska that yielded mature ascospores was anomalous in that part of the collection lacked atranorin (by TLC and lack of crystals in the cortex), the first such specimen recorded. Some ascospores were of the Physcia-type but many clearly trended towards the Physconia-type, although retaining some apical wall thickening at maturity. The small sample of ascospores measured from Alaskan material, (18·5–)22·0–24·0(–24·5) × (10·0–)10·5–12·5(–13·0) µm (n=19), l/w ratio (1·7–)1·8–2·1(–2·2), was comparable with the Japanese material cited above. All mature ascospores had a prominent torus but most had unusually light ornamentation. Another fertile, sorediate specimen from Alaska, which was not subject to molecular analysis, had normal chemistry with atranorin and zeorin present.
The Scandinavian and non-Alaskan western North American specimens (pamk cluster 2; Fig. 2) were sterile. However, ascospore sizes for Rinodina degeliana quoted by Mayrhofer & Moberg (Reference Mayrhofer and Moberg2002) are comparable with those given above. The very complete type description (Coppins Reference Coppins1983) agrees with our observations of the holo- and isotype except that the ascospores, when fully developed, belong to the Physconia-type rather than the Dirinaria-type of the protologue, or the Physcia-type as reported by Mayrhofer & Moberg (Reference Mayrhofer and Moberg2002) and Sheard (Reference Sheard2010). Coppins (Reference Coppins1983) reported ascospore measurements of 19–25×10–14 µm that were confirmed by our observations (20·5–25·0×10·5–13·0 µm, n=14) of the type specimens.
We found that specimens from all clades may possess soredia. Soredia were measured under the compound microscope to an accuracy of 5 µm but showed only a limited variation in size. Specimens from Norway and Washington (P102 and P233 respectively, pamk cluster 2) developed slightly smaller (median 25 µm diam.) soredia compared to 30 µm in other regions of the world ((20–)25–30(–40) µm, n=115). However, this small size difference is accommodated in the 25–75 of the above percentile range and is therefore not meaningful. Soralia develop marginally (somewhat labriform; Fig. 3F & G) on the areoles, and are of similar size in the different clades.
Concerning Rinodina mniaraea, Sheard (Reference Sheard2010) noted that he searched in vain for morphological, anatomical and ascospore characters to separate the different chemotypes. Numerous collections by Imshaug (MSC) in the southern Rocky Mountains offered the opportunity to test ascospore sizes of different chemotypes from the same collections or from the same localities. Some significant differences were found but they were not consistent between the chemotypes. Therefore, Sheard (Reference Sheard2010) was unable to recognize the chemical variants as being taxonomically distinct. Descriptions of the morphology of the taxa based on the chemical variants are found in the following section.
Taxonomy of Rinodina subparieta
Rinodina subparieta (Nyl.) Zahlbr.
MycoBank No.: MB816978
Cat. Lich. Univers. 7: 554 (1931).
Basionym: Lecanora subparieta Nyl., Acta Soc. Sci. Fenn. 26: 30 (1900); type: Japan, Itchigômé, 1879, Almquist (H-Nyl 28856––holotype!).
Rinodina degeliana Coppins, Lichenologist 15: 147 (1983); type: Sweden, Lule Lappmark, Kvikkjokk parish, south-east slope of Nammatj, 66°56'N, 17°42'E, alt. 520 m, on Salix bark in Picea woodlands, 1977, Coppins 6238 & Tibell (E––holotype!; UPS––isotype!).
Thallus epiphloeodal, thin, light grey, areoles to 0·50–0·80 mm wide, subangular or with minutely sublobate margins; areoles isolated around margin, rarely coalescing to become rimose-areolate; surface plane, sometimes becoming rugose, matt or somewhat shining; margin determinate; prothallus absent or very thin, sometimes black, visible between marginal areoles; soredia whitish (paler than thallus), 20–40 µm diam., arising from underside of slightly upturned areole margins to form labriform soralia (Fig. 3F & G), areoles sometimes dissolving into diffuse soralia, or soredia rarely absent.
Apothecia frequently absent, when present typically 1 per areole, becoming narrowly attached, numerous and often contiguous, to 0·50–0·90 mm diam. (Fig. 3A–C); discs dark brown or typically black, plane sometimes becoming convex, rarely half-globose; thalline margin 0·05–0·10 mm wide, entire (rarely sorediate in part), persistent or becoming excluded; excipular ring absent or visible when apothecia moistened, narrow and confluent. Thalline exciple 60–100 µm wide laterally; cortex 10–20 µm wide; sometimes with epinecral layer 5–10 µm wide; crystals in cortex (atranorin), sometimes in medulla (zeorin); cortical cells to 3·5–6·0 µm, not pigmented, mostly hidden by crystals; algal cells to 6·0–12·0 µm wide; thalline exciple expanding to 90–140 µm below, cortex 25–70 µm deep; proper exciple hyaline or very light brown, 5–10 µm wide laterally, expanding to 20–30 µm wide above; hypothecium hyaline to very light brown, 70–140 µm deep; hymenium (75–)90–120 µm high, not inspersed; paraphyses 2·0–2·5 µm wide, strongly conglutinate, apices 3·5–4·5 µm wide, brown capitate, immersed in dispersed pigment forming a red-brown epihymenium; asci 60–80×15–30 µm. Ascospores (rarely 4–)8 per ascus, type A development, Physcia- to Physconia-type (Fig. 3D & E), (18·0–)20·0–23·5(–25·5)× (10·0–)10·5–12·0(–13·0) µm (n=205), l/w ratio (1·6–)1·8–2·1(–2·2), thickened apical walls during early development and frequently arrested at this stage; torus present; walls mostly darkly pigmented, typically strongly ornamented (×1250).
Pycnidia rare, c. 0·05 mm diam., immersed in thallus, ostioles brown to black; pycnoconidia hyaline, bacilliform 3·5–4·5 ×1·0 µm.
Chemistry. Spot tests K+ yellow, C−, P+ bright lemon yellow (very rarely negative). Secondary metabolites: atranorin (very rarely absent) and zeorin. Cortical atranorin typically abundant as indicated by strength of P+ spot test.
Ecology. Corticolous, mainly on deciduous trees.
Notes. The revised description provided here is based on our studies of Rinodina degeliana from Europe (Coppins Reference Coppins1983; Tønsberg Reference Tønsberg1992; Mayrhofer & Moberg Reference Mayrhofer and Moberg2002), North America (Sheard Reference Sheard2010) (with the exception of the ascospore dimensions), the type of R. subparieta and fertile specimens from Japan collected by GT and TT. Rinodina subparieta is a variable species. In addition to having fertile sorediate, fertile esorediate (rare) and non-fertile sorediate morphs (the most frequent state), ascospore size and the development stage attained are also variable. Such ascospore variability is typical of many Rinodina species possessing vegetative diaspores (J. W. Sheard, pers. obs.).
The very detailed protologue of Rinodina degeliana (Coppins Reference Coppins1983) agrees with our observations of the types of that species. We found only one important difference: that fully developed ascospores belong to the Physconia-type, rather than the Dirinaria-type or Physcia-type as reported by Mayrhofer & Moberg (Reference Mayrhofer and Moberg2002) and Sheard (Reference Sheard2010), respectively. Sheard (Reference Sheard2010) questioned the distinction between the Physcia- and Physconia-ascospore types, citing the many Rinodina species that have intermediate ascospore types. Rinodina subparieta is another such example, with specimens exhibiting the Physcia-spore type having arrested ascospore development, a very common feature of the species in Europe, North America and Scandinavia. The ascospore dimensions cited above are from the total of specimens examined in the present study.
The species is easily distinguished from other sorediate Rinodina species (such as R. efflorescens, R. griseosoralifera Coppins, R. sheardii Tønsberg and R. stictica Sheard & Tønsberg) by its typically light grey areoles with marginal, labriform soralia and whitish soredia, and chemically by the presence of abundant cortical atranorin (P+ strong lemon yellow) and zeorin in its medulla. Very rarely the thallus and the soredia may have a greenish tinge (e.g. specimen in Fig. 3A). We have noted two thalline morphotypes. Most common is the morph with a plane and somewhat shining (waxy) surface, the areoles more or less angular and 0·6–0·8 mm wide, to which the type specimens of R. degeliana belong. Very rarely such areoles coalesce to form a rimose-areolate thallus. The second morph, which may be more common in fertile specimens, has smaller areoles, rarely exceeding 0·5 mm in width, with a matt, slightly convex surface and minutely sublobate margins before breaking into soredia. It is notable that the esorediate form of R. subparieta has only been found at very high elevations on Honshu, Japan (1960–2550 m), whereas sorediate forms, widely distributed geographically in the Northern Hemisphere, are found at lower elevations, although at up to 1725 m on Honshu (J. W. Sheard, pers. obs.), and only at high elevations in the southern Appalachians (1502–1920 m, Lendemer 2528, 44770, 43873 NY).
Additional fertile, esorediate specimens examined. Japan: Honshu: Etchu Prov., Nakasinkawa-gun, 33 km ESE Toyama, Midagahara, along path to Tateyama crater 36°34'N, 137°34'E, alt. 1960–2000 m, subalpine open mixed deciduous Abies mariesii forest, on dead, standing, exposed Abies mariesii, 1994, Thor 12708 (UPS–duplicate in TNS, not seen); Kai Prov. (Yamanashi Pref.), Kofu City, along trail between Odarumi Pass and Mt. Kinpu (Kimpō), old-growth, subalpine forest dominated by Abies mariesii and Tsuga diversifolia, on Abies mariesii, alt. 2552 m, 35°52·163'N, 138°37·930'E (WGS84, ±10 m), 2012, Thor 27994 (UPS); Makioka-cho, Yamanashi City, Odarumi pass, near mountain hut, old-growth, subalpine forest dominated by Abies mariesii, on A. mariesii, alt. 2368 m, 35°52·171'N, 138°40·001'E (WGS84, ±100 m), 2012, Thor 28096 (UPS); on Sorbus sp., Thor 28120 (UPS).
Selected sorediate specimens examined. Japan: Hokkaido: Ishikari Prov., Kamikawa-gun, Kamikawa-cho, 0·5–1·5 km E of Obako Gorge tourist centre, along gravel road just N of Niseicharomappu-gawa stream, open and plane land with deciduous trees, on Abies sachalinensis, alt. 720 m, 43°42'N, 143°01'E, 1995, Thor 14624 (UPS); Kitami Prov., Esashi-gun, Esashi-cho, 2 km S of Honcho, just SW of road 238, plantation of Abies sachalinensis and Larix gmelinii var. japonica with scattered Quercus mongolica var. grosseserrata, on Larix, alt. 10–20 m, 44°55'N, 142°35'E, 1995, Thor 14251 (UPS); Roshiri Island, Oshidomari area, along the path from Rishiri-hokuroku campsite (2·5 km S of Sakae Town) to Mt. Pon (near the campsite), 45°13'N, 141°13'E, alt. 220 m, old-growth humid forest dominated by Abies sachalinensis, corticolous on Abies sachalinensis, 1995, Tønsberg 22289 (BG); Shari-gun, Shari-cho, Shiretoko National Park, 9 km NE of Utoro Village, along trail around Shiretoko-goko Lakes, 44°07'N, 145°05'E, alt. 260 m, old-growth, mixed deciduous/coniferous forest, on Betula, 1995, Tønsberg 22797 (BG); Kushiro Prov., Akkeshi-gun, Akkeshi-cho, 44 km E of Kushiro City, S Akkeshiko Lake, just S of road following coast, humid mixed Abies sachalinensis/deciduous forest, on Quercus mongolica var. grosseserrata, alt. 120 m, 42°60'N, 144°57'E, 1995, Thor 14486 (UPS); Hamanaka-cho, 55 km E of Kushiro City, 3 km E of Hichiripputo Lake, just S of road following coast, 43°02'N, 145°03'E, alt. 60 m, humid, mixed Abies sachalinensis/deciduous forest, corticolous on Sorbus, 1995, Tønsberg 22936b (BG); Nemuro Prov., Shiretoko Peninsula, Menashi-gun, Rausu-cho, just S of road 334 crossing Shiretoko Peninsula, 4 km WNW of Sakae City, Rausu hot spring, open deciduous forest, on Alnus sp., alt. 130 m, 44°02'N, 145°09'E, 1995, Thor 14406 (UPS); Teshio Prov., Rumoi-gun, Obira-cho, 21 km ENE of small town Obira at the coast, along the trail from parking area to Tengunotaki Waterfall, 44°04'N, 141°55'E, alt. 50–100 m, dense, old-growth, mixed deciduous forest with scattered Abies sachalinensis, corticolous on Betula, 1995, Tønsberg 21955 (BG); Tokachi Prov., Kato-gun, Kamishihoro-cho, 6 km S of Mikumi Tunnel through Mt. Mikumi-yama, just W of road 273, marsh with Abies sachalinensis/deciduous forest, on dead deciduous tree, alt. 680 m, 43°32'N, 143°09'E, 1995, Thor 14581 (UPS). Honshu: Bizen Prov. (Okayama Pref.), Maniwa-gun, Kawakami-mura, 1–2·3 km N of Hiruzen Research Institute, along road, on exposed tree at roadside, alt. 500–550 m, 35°18'N, 133°38'E, 1994, Thor 12212 (UPS); Etchu Prov. (Toyama Pref.), Nakashinkawa-gun (Nakaniikawa-gun), Tateyama-cho, 25 km ESE of Toyama, Bijodaira, path S of Bijodaira bus stop, mixed deciduous/Cryptomeria japonica forest, on old deciduous tree, alt. 960–980 m, 36°35'N, 137°28'E, 1994, Thor 12664 (UPS); Kai Prov. (Yamanashi Pref.), Makioka-cho, Yamanashi City, at road to Odarumi pass, open forest dominated by Larix, on Prunus sp., alt. 1726 m, 35°49·172'N, 138°38·963'E (WGS84, ±30 m), 2012, Thor 28169 (UPS); Shimotsuke Prov. (Tochigi Pref.), 20 km WNW of Nikko, 1 km S of Yumoto Village (36°48'N, 139°26'E), Yudaki Falls, on deciduous tree in open, mixed forest, alt. 1440–1480 m, 1994, Thor 12771 (UPS).
Taxonomy of Rinodina mniaraea s. lat.
Rinodina cinnamomea (Th. Fr.) Räsänen
Ann. Acad. Scient. Fenn. 34(4): 137 (1931).
Basionym: Rinodina mniaraea var. cinnamomea Th. Fr., Nova Acta Regiae Soc. Sci. Upsal. 3(3): 228 (1860); type: Norway, Finnmark, Syd-Varanger, Pasvig [Pasvik], 1857, Th. Fries (UPS––lectotype! designated by Mayrhofer & Moberg Reference Mayrhofer and Moberg2002: 72).
Rinodina mniaraea var. chrysopasta (Lettau) Zahlbr., Cat. Lich. Univ. 7: 533 (1931).—Rinodina mniaraea f. chrysopasta Lettau, Hedwigia 60: 123 (1918); type: Switzerland, Engadin, am Morteratschgletscher bei Pontresina zwischen Gletscherzunge und dem Gasthaus, 1900 m, 1912, Lettau (B––holotype!).
Thallus thick, brown, dark brown to copper brown, continuous, rugose, matt.
Apothecia abundant, broadly attached, 0·3–1·5 mm diam.; disc dark brown to black-brown, rarely pruinose, plane to convex to strongly convex; thalline exciple 30–80 µm wide laterally; cortex indistinct; epinecral layer frequent, 10–30 µm wide; hypothecium hyaline or light brown at the base, 20–200 µm deep, inspersed with oil droplets; hymenium 80–130 µm high; epihymenium red-brown to brown. Ascospores Physcia-type, 19·0–27·5–35·0×10·0–12·5–15·0 µm, average l/w ratio 2·2.
Pycnidia not observed.
Chemistry. Orange to yellow pigment especially in the lower part of the medulla reacting K+ purple to violet; chemotype A: Skyrin, ±variolaric acid, ±cinnamomea unknown (5/3/5), ±atranorin; chemotype B: 1-O-methylemodin or 8-O-methylemodin, ±variolaric acid, ±cinnamomea unknown, ±atranorin.
Ecology and distribution. On soil, bryophytes and decaying plants often above slightly calcareous and acid bedrock in arctic-alpine habitats. Widespread across the Northern Hemisphere, scattered in more southerly mountains such as the Rocky Mountains, northern Scotland, Alps, Carpathian Mountains, Balkan Peninsula, Anatolian Mountains, Caucasus, Tibet, Himalayas (Mayrhofer & Moberg Reference Mayrhofer and Moberg2002; Strasser et al. Reference Strasser, Hafellner, Stešević, Geci and Mayrhofer2015).
Notes. This species is characterized by a yellow to orange medulla. Chemotype A possesses the anthraquinone skyrin and chemotype B the anthraquinone 1-O-methylemodin or 8-O-methylemodin. Additional chemical compounds that may occur in different amounts are atranorin, variolaric acid and cinnamomea unknown. Based partly on the occurrence of atranorin, Mayrhofer & Moberg (Reference Mayrhofer and Moberg2002) distinguished a total of four chemotypes. The type specimens of R. mniaraea var. cinnamomea and R. mniaraea var. chrysopasta belong to chemotype A.
Selected specimens examined. France: Dept. Hautes-Alpes: Dauphine, Umgebung des Col du Lautaret, Epaulement central de Combeynot, 2200–2600 m, 1957, Poelt 878 (GZU).—Germany: Bavaria: Allgäuer Alpen, Höfats-Gufel, 2000 m, 1918, Lettau (B).—Montenegro/Kosovo: Prokletije: Hajla, summit of Hajla mountain, 2403 m, 2010, Mayrhofer 464 & Stešević (GZU).—Norway: Finnmark: Mageröy, Skarvaag, 1864, Th. Fries (UPS).—Switzerland: Graubünden: Val Fenga, Ritzenjoch, 2500 m, 1967, Poelt & Vězda (BRA).—China: Tibet: Prov. Xizang, Ningjing Shan Mts., 9 km W Markam (=Gartog), c. 4000 m, 1994, Obermayer 3798 (GZU).—Georgia: Dzhava: Kaukasus montes Rachinski khrebet, in vicinitate pagi Tschordi, in monte Tagverula, 2300–2600 m, 1989, Vašak (GZU).—Nepal: Mahalangur Himal: Khumbu, Höhe westlich über Gorak Shep, 5300–5400 m, 1962, Poelt L905 (M).—Pakistan: W-Himalaya: Kaghan Valley, above Saiful-Muluk-Lake, 4070 m, 1990, Schickhoff 102/f4 (GZU).—Turkey: Provinz Bursa: Ulu Dag, 20 km SE von Bursa, 2150 m, 1976, Kalb 16068 & Plöbst (WIS).
Rinodina mniaraea (Ach.) Körb.
Syst. Lich. Germ.: 126 (1855).
Basionym: Lecanora “mniaroea” Ach., Syn. Lich.: 339 (1814); type: [Switzerland], Helvetia, Schleicher (H-ACH 1136 “91a”––lectotype! designated by Mayrhofer & Moberg Reference Mayrhofer and Moberg2002).
Rinodina mniaroeoides (Nyl.) H. Olivier, Mém. Soc. Sci. Nat. Cherbourg 37: 162 (1909).—Lecidea mniaroeoides Nyl., Flora 53: 36 (1870); type: [Russia], Lapponia Kemensis, Muonio, Olostunturi, 1867, Norrlin (H-NYL 10657—lectotype! here designated).
Rinodina amniocola (Ach.) Körb., Parerg. Lich.: 73 (1859).—Lecanora amniocola Ach., Syn. Lich.: 156 (1814); type: [Switzerland], Helvetia, Schleicher (H-ACH 1187––holotype!).
Rinodina turfacea var. nuda Körb., Parerg. Lich.: 72 (1859); type: [Switzerland], Auf der Erde zwischen Moosen bei St. Moritz, Hepp [Hepp: Die Flechten Europas Nr. 83] (G––lectotype! here designated).
Note. Lecanora amniocola was described in the same publication as Lecanora mniaroea, the latter in the appendix. The index was given after the appendix, meaning that it was published at the same time in one issue. We therefore retain the name mniaraea, which was used earlier in Rinodina.
(Fig. 3H)
Thallus usually well developed, grey-brown to brown, continuous, surface usually rugose, rarely developing raised verrucae, matt.
Apothecia broadly attached and often contiguous, 0·7–1·1 mm diam.; disc dark brown to black-brown, sometimes pruinose especially when young, often becoming convex; thalline exciple 70–100 µm wide laterally; cortex 5–10 µm wide; epinecral layer frequent, 5–10 µm wide; hypothecium hyaline or light brown in specimens with dark thalli, 80–250 µm deep, inspersed with oil droplets; hymenium 80–130 µm high; epihymenium red-brown. Ascospores Physcia-type 21·0–27·5–35·0×10·0–12·5–15·0 µm, average l/w ratio 2·2.
Pycnidia immersed in thallus; pycnoconidia bacilliform, 4·0−5·0×1·0 µm.
Chemistry. Spot tests all negative, secondary metabolite±variolaric acid.
Ecology and distribution. On soil, bryophytes and decaying plants above calcareous and acid bedrock in arctic-alpine habitats, rarely descending into the subalpine forest zone. Widespread across the Northern Hemisphere, scattered in more southerly mountains such as the Rocky Mountains, Sierra Nevada, Pyrenees, Alps, Sudeten, Carpathians, Balkan Peninsula, Caucasus, central Asia, Karakorum and Himalayas (Mayrhofer & Moberg Reference Mayrhofer and Moberg2002; Mayrhofer et al. Reference Mayrhofer, Denchew, Stoykov and Nikolova2005; Sheard Reference Sheard2010). The record from the South Shetland Islands (Antarctica) by Olech (Reference Olech1989) needs confirmation.
Note. Rinodina mniaraea is characterized by the lack of other secondary metabolites apart from variolaric acid.
Selected specimens examined. Germany: Allgäuer Alpen: Obermädli Alpe, Rehm (W). Bayrische Alpen: Chiemgau, Hochfelln und Hochgern, Britzelmayr (PRM).—Switzerland: Bern: Oberaaralp–Bärenegg, 2400 m, 1920, Frey 9075 (G). Graubünden: Val Fenga, ad latera montis Heidelberger Spitze, 2500 m, 1967, Poelt & Vězda (BRA). Wallis: Zermatt, Gornergrat, Hochtälligrat, 3130–3150 m, 1961, Frey 27168 (G).—Georgia: Caucasus Magnus: region montis Elbrus, ad latera austro-occidentalia montis Čeget, supra vallem torrentis Baksan, 2800 m, 1980, Vězda 20659 (PRM).—Italy: South Tyrol: Hohe Tauern, Venediger Gruppe, Rieserferner-Ahrn Natur Park, SE of Kasern, Röttal, glacier forefield of the Rötkees, 2345 m, 2013, Bilovitz, Nascimbene, Tutzer, Wallner & Mayrhofer (GZU).—Mongolia: Archangai Aimak: Changai, Tarbagatai, 5 km vom Solon-Got-Paß, 2400 m, 1983, Huneck (GZU).—Nepal: Zentral-Himalaya: Langtang-Gebiet, Yala, oberes Langtang, 4830 m, 1986, Miehe & Miehe 4595 (GZU).—Pakistan: Karakorum: Shinghai Gali, 4580 m, 1990, Miehe & Miehe 1190 (GZU).—Turkey: Provinz Bursa: Ulu Dag, 20 km SE von Bursa, 2150 m, 1976, Kalb 15879, 16061, 16063 & Plöbst (WIS).
Rinodina mniaraeiza (Nyl.) Arnold
Flora 53: 469 (1870).
Basionym: Lecanora “mniaroeiza” Nyl., Flora 53: 33 (1870); type: [Finland], Tavastia australis, Häme, Padasjoki, 1866, Norrlin (H-NYL 28734––lectotype! designated by Mayrhofer & Moberg Reference Mayrhofer and Moberg2002).
Rinodina mniaraea f. biatorina (Nyl.) Arnold, Verh. Zool.-Bot. Ges. Wien 37: 132 (1887).—Lecanora turfacea var. biatorina Nyl., Lich. Scand.: 151 (1861); type: Norway, Sør-Trøndelag, Dovre, Schimper (H-NYL 28717––lectotype! here designated).
Rinodina hookeri (Fr.) Dalla Torre & Sarnthein, Flecht. Tirol : 206 (1902).—Parmelia hookeri sensu Fr. non Lichen hookeri Borr. ex Sm., E. Fries, Lich. Eur.: 94 (1831); type: [Switzerland], Helvetia, Schleicher (UPS––lectotype!, here designated).
Thallus light grey, rarely pale brown to brown, continuous, rugose and matt.
Apothecia immersed to broadly attached, 0·6–0·9 mm diam.; thalline exciple 35–70 µm wide laterally; cortex indistinct; disc dark brown to black-brown, without pruina, plane to convex; hypothecium 70–250 µm deep, hyaline or light brown to dark brown at the base, inspersed with oil droplets; hymenium 85–150 µm high; epihymenium light brown. Ascospores Physcia-type, 20·0–25·0–31·5×9·0–11·5–15·0 µm, average l/w ratio 2·15.
Pycnidia immersed; pycnoconidia bacilliform, 3·8–4·5 × 1·0 µm.
Chemistry. Spot test K+ yellow, secondary metabolites atranorin, ±variolaric acid.
Ecology and distribution. On soil, bryophytes and decaying plants above calcareous and more rarely acid bedrock in arctic-alpine habitats. Widespread across the Northern Hemisphere, scattered in more southerly mountains such as the Rocky Mountains, Northern Scotland, Alps, Carpathians, Balkan Peninsula, Anatolian mountains and Karakorum (Mayrhofer & Moberg Reference Mayrhofer and Moberg2002; Strasser et al. Reference Strasser, Hafellner, Stešević, Geci and Mayrhofer2015).
Note. Rinodina mniaraeiza is characterized by the presence of atranorin and can be distinguished from R. mniaraea and R. cinnamomea by the lighter thalli and by slightly smaller ascospores.
Selected specimens examined. Austria: Niederösterreich: Lunz am See, Scheiblingstein, 1500–1600m, 1995, Türk 20285 (GZU).—Germany: Bavaria: Wettersteingebirge, Hänge nördlich der Alpspitze, c. 2000 m, 1966, Poelt 13457 (GZU).—Switzerland: Bern: Grimsel, without date and collector, (BP); Fribourg, Simmental, Kaiseregg, östlicher Vorgipfel ob Waloalp, 2050 m, 1926, Frey 10010 (G). Graubünden: Unterengadin, Nationalpark, Buffalora Alp, 2140 m, Frey 10037 (G). Obwalden: Frutt, in den Schratten, 1920 m, 1921, Frey 28202 (G).—Pakistan: Karakorum: Baltistan, Haramosh Range, Alm Pakora SE Ganto La, 3700–3900 m, 1991, Poelt K91-171 (GZU). NW-Himalaya: Eastern Dersai Plateau, 3950 m, 1991, Poelt K91-267 (GZU).—Turkey: Provinz Bursa: Ulu Dag, 20 km SE von Bursa, 2150 m, 1976, Kalb 16048 & Plöbst (WIS).
Discussion
In the present study we have investigated the status of Rinodina degeliana, R. subparieta and R. mniaraea s. lat. By evaluating evidence derived from molecular phylogenetic, chemical and anatomical studies we provide hypotheses about the taxonomy and updated nomenclature for these crustose lichen lineages.
The status of Rinodina subparieta and R. degeliana
We recovered samples of Rinodina degeliana and R. subparieta in two well-supported clades in our concatenated maximum likelihood phylogeny (Fig. 1A). According to pamk-clustering based on the bGMYC results (Fig. 2), the two Rinodina subparieta samples were assigned to cluster 1 together with samples of R. degeliana. However, species delimitation also recovered Rinodina degeliana in two additional clusters (pamk cluster 2–3; Fig. 2), indicating multiple previously undiscovered lineages in R. degeliana.
The only reliable morphological character to separate Rinodina degeliana and R. subparieta was the lack of soralia in R. subparieta. We took an a posteriori approach to identify other possible synapomorphies informed by our phylogenetic hypothesis. Our morphological investigations, however, did not reveal any reliable differences between the R. degeliana clades or between R. degeliana and R. subparieta.
Fertile specimens were rarely observed in the material investigated and occasionally they lacked soralia. Ascospores from this material are variable in size and structure, even when immature and overmature ascospores were excluded from measurement. Such variable ascospores are typical for Rinodina species that tend to rely on vegetative reproduction (J. W. Sheard, pers. obs.). Young, not fully pigmented ascospores have Physcia-type lumina and mature into darkly pigmented Physcia-type ascospores or into Physconia-type ascospores also with darkly pigmented walls, a prominent torus and with rather heavily ornamented walls, although the latter character is sometimes less well developed. Asynchronous ascospore development was also frequent among specimens with Physconia-type spores; rarely only four ascospores in the ascus reach maturity.
The frequent development of Physcia-type ascospores into Physconia-type would seem to confirm the opinion of Sheard (Reference Sheard2010) that these ascospore types should be regarded as developmental phases of a single ascospore type. Both are accompanied by a distinctive red-brown epihymenium colour in all clades, as are other species possessing these ascospore types (Ropin & Mayrhofer Reference Ropin and Mayrhofer1993). This coloration is derived from a dispersed red-brown pigment in the epihymenial gelatin together with the dark brown or black capitate pigmentation of the terminal cells of the paraphyses.
Conclusively, R. degeliana and R. subparieta (and also the different R. degeliana clades) were indistinguishable in respect to morphology, anatomy and chemistry. However, molecular results indicate multiple, although in part poorly supported, clades and clusters (Figs. 1A & 2). This could imply multiple species-level lineages in Rinodina degeliana. The specimens of R. subparieta available to us were nested in one of the R. degeliana clades and form a cluster with R. degeliana ITS sequences in bGMYC. Our molecular results and the lack of phenotypic characters to separate the two species thus lead us to conclude that R. subparieta and R. degeliana are conspecific.
We recognize that our interpretation of this species is broad and also includes specimens assigned to possible species-level lineages containing only R. degeliana specimens (pamk clusters 2 & 3; Fig. 2). This may represent an oversimplification of the taxonomic status of these lineages. At this point, the formal introduction of new names for any of the three clades would have to be based on molecular evidence only. It is generally possible to describe species based mainly on clade affinity in phylogenetic trees (Leavitt et al. 2013 Reference Leavitt, Fernández-Mendoza, Pérez-Ortega, Sohrabi, Divakar, Lumbsch and St. Claira ), however we refrain from doing so in the case of Rinodina degeliana/R. subparieta.
Another solution to this problem, although probably impossible, would be to sequence the type specimens of R. subparieta and R. degeliana, ascertain which names apply to individual clades and adjust taxonomy accordingly. Since R. subparieta is nested in a clade (Fig. 1A) and cluster (Fig. 2) with R. degeliana samples and we lack morphological, anatomical and chemical characters to distinguish these lineages, we have been conservative in our interpretation. Further studies, particularly in view of a possible geographical separation, may help reveal the evolutionary history of the clades. Until additional data are available, we propose to use the name R. subparieta, which takes priority over R. degeliana.
The status of R. mniaraea s. lat.
We recovered Rinodina mniaraea in three clades (Fig. 1A) and detected three potential species-level lineages in the pamk clustering analysis (Fig. 2). The three clusters correspond to the three previously recognized varieties Rinodina mniaraea var. cinnamomea, R. mniaraea var. mniaraeiza and R. mniaraea var. mniaraea. Searches for morphological and anatomical differences between these lineages have been unsuccessful (Sheard Reference Sheard2010). All form Lecanora-type asci with eight Physcia-type ascospores, (20·5–)24·5–25·5(–30·0)×(9·5–)11·5–12·5(–14·5) µm, that have thick apical walls and septal wall thickenings separating the usually unequal-sized cells (Sheard Reference Sheard2010).
Differences between R. mniaraea var. cinnamomea, R. mniaraea var. mniaraeiza and R. mniaraea var. mniaraea are restricted to secondary chemistry (Mayrhofer & Moberg Reference Mayrhofer and Moberg2002): var. mniaraea possesses either no secondary compounds or variolaric acid while var. mniaraeiza possesses atranorin and sometimes variolaric acid; var. cinnamomea produces the anthraquinones skyrin and 1-O-methylemodin or 8-O-methylemodin, sometimes with atranorin and variolaric acid or both, and one additional unknown.
Räsänen (Reference Räsänen1931) had already established R. cinnamomea (Th. Fr.) Räsänen and Arnold (Reference Arnold1870) transferred Lecanora “mniaroeiza” described by Nylander (Reference Nylander1870) to Rinodina. It remains unclear if the lineages discovered by us are truly distinct species. We cannot rule out the alternative hypothesis that R. mniaraea varieties all belong to a single species. In the mtSSU gene tree (Fig. 1C) Rinodina mniaraeiza does not form a monophyletic group and we thus detected a topological conflict between gene trees. A topological conflict could indicate gene flow. However, at present the size of our molecular dataset of R. mniararea s. lat. prevents us from investigating this issue in detail. An alternative explanation could be recent speciation with still incomplete lineage sorting. Additional data are needed to resolve this issue. We failed to find anatomical or morphological characters to separate taxa in R. mniarea s. lat. in a situation similar to that of R. subparieta/R. degeliana. Secondary chemistry, however, is characteristic and corresponded to clade (Fig. 1A) and cluster (Fig. 2) affinity in all R. mniaraea s. lat. specimens in our dataset. This is similar to other studies where morphologically indistinguishable taxa were proposed to be chemically distinct species (Lendemer Reference Lendemer2012; Leavitt et al. 2013 Reference Leavitt, Esslinger, Nelsen and Lumbschb ). Our consistent chemical, phylogenetic (but see mtSSU gene tree; Fig. 1C) and species delimitation results lead us to follow Räsänen and Arnold and propose that the varieties of R. mniaraea s. lat. should be recognized at the species level.
The placement of Oxnerella safavidiorum in Physciaceae
The monotypic genus Oxnerella was recently described from the Iranian province Esfahan (Kondratyuk et al. 2014 Reference Kondratyuk, Lőkös, Kim, Kondratiuk, Jeong, Zarei-Darki and Hurb ). Oxnerella safavidiorum possesses a crustose rusty brown thallus, lecanorine apothecia with a Biatora-type ascus and hyaline 1-septate ascospores (Kondratyuk et al. 2014 Reference Kondratyuk, Lőkös, Kim, Kondratiuk, Jeong, Zarei-Darki and Hurb ). Kondratyuk et al. (2014 Reference Kondratyuk, Lőkös, Kim, Kondratiuk, Jeong, Zarei-Darki and Hurb ) conducted a phylogenetic analysis based on ITS and mtSSU sequences but could not resolve the placement of Oxnerella. A possible explanation is that because Oxnerella possesses hyaline 1-septate ascospores, Kontratyuk et al. (2014b) assumed a position close to Lecania in the family Ramalinaceae.
Our phylogenetic analysis now resolves Oxnerella safavidiorum as sister to Rinodina bischoffii with high support (82% BS; Fig. 1A). This placement should be considered tentative as long as fresh material is unavailable to verify sequence identity and rule out contaminations of Oxnerella type material. The differences in ascus type and ascospores of Oxnerella and Rinodina are surprising. A verification of ascus and ascospore characters of Oxnerella also has to be postponed until additional material for study is available to us.
We thank Fernando Fernández-Mendoza for valuable discussions about the species delimitation analyses, Peter Kosnik for support with TLC, the curators of the herbaria B, BP, BRA, E, G, GZU, H, PRM, UPS and W, and Klaus Kalb for the loan of specimens. The herbarium of Klaus Kalb was recently deposited in WIS. We thank Peter Bilovitz for providing the image of R. mniaraea and James Lendemer for supplying a fresh collection for molecular analysis. Funding for this project comes from the Austrian Science Fund (FWF) in the projects P25078-B16 and P25237-B16. TT also thanks Bruce McCune for the invitation to do lichenological fieldwork in Alaska, the National Park Service (NPS), Southwest Alaska Network, Anchorage, for funding, and Amy Miller and James Walton (both NPS) for project coordination and for organizing and executing field logistics. We thank Jiří Malíček for bringing the Oxnerella problem to our attention. François Lutzoni and James Lendemer, and the anonymous reviewers, are thanked for valuable comments on the manuscript.







