Multidrug resistance (MDR) among Enterobacteriaceae has increased worldwide since 2000, with resistance to extended-spectrum cephalosporin and carbapenem antibiotics emerging as specific threats.Reference Canton, Coque and Baquero 1 Outbreaks of MDR gram-negative bacteria in hospitals and long-term-care facilities are well documented, and nosocomial infections with such organisms are increasing.Reference Canton, Coque and Baquero 1 , Reference McGrath and Asmar 2 Reported vehicles of transmission include environmental sources (eg, ventilators, patient beds, infusion pumps, sink drains)Reference Palmore and Henderson 3 – Reference Lerner, Adler, Abu-Hanna, Meitus, Navon-Venezia and Carmeli 5 and hands of healthcare workers.Reference Hornbeck, Naylor, Segre, Thomas, Herman and Polgreen 6 , Reference Pittet, Allegranzi and Boyce 7 Outbreaks have been linked to medical devices and procedures,Reference Hidron, Edwards and Patel 8 , Reference Emori and Gaynes 9 including catheters,Reference Wenzel, Osterman and Donowitz 10 bronchoscopes,Reference DiazGranados, Jones and Kongphet-Tran 11 and endoscopesReference Gastmeier and Vonberg 12 —particularly those used in endoscopic retrograde cholangiopancreatography (ERCP).Reference Coelho-Prabhu, Shah, Van Houten, Kamath and Baron 13 – Reference Epstein, Hunter and Arwady 16
In October 2012, Washington State began surveillance for carbapenem-resistant (CR) Enterobacteriaceae through a voluntary reporting mechanism after cases had been identified in previous years. The Washington State Public Health Laboratory conducts phenotypic testing on submitted isolates, and all confirmed CR isolates are subjected to polymerase chain reaction (PCR) testing to identify 5 common carbapenemase genes (bla KPC, bla NDM, bla IMP, bla VIM, and bla OXA-48). PCR-negative isolates are subjected to a secondary PCR panel to identify common extended-spectrum cephalosporinases (bla CTX-M and bla CMY).
In early 2013, we identified a cluster of 3 carbapenemase PCR-negative, CR Escherichia coli isolates that shared a novel bla CMY-2 allele and a distinctive fumC/fimH typing profile. These genetic markers were unique and had not been identified previously in Washington or in the reported literature, suggesting the isolates had a common source. No further isolates were identified until mid-2013, and by September 2013, a total of 7 matching isolates had been identified. Initial review of the 7 patients’ medical histories revealed that all patients had complicated pancreatic or biliary disease and had undergone ERCP procedures at hospital A. Additionally, 3 patients had a phenotypic AmpC E. coli (defined as cefoxitin and third-generation cephalosporin resistant, but carbapenem sensitive) recovered weeks before isolation of CR E. coli. Of note, an increase in phenotypic AmpC E. coli had been observed at hospital A before this outbreak of CR E. coli isolates.
In collaboration with hospital A staff, we conducted a public health investigation to determine the extent and epidemiologic characteristics of the outbreak, identify potential sources of transmission, design and implement infection control measures, and determine the association between the CR E. coli and AmpC E. coli circulating at hospital A.
METHODS
Definitions
The outbreak strain was defined as including AmpC E. coli isolates from patients cared for at hospital A starting in January 2012 and either (1) indistinguishable, closely related, or possibly related by pulsed-field gel electrophoresis (PFGE), or (2) containing identical bla CMY-2, fumC, and fimH alleles regardless of PFGE result. Cases were defined as hospital A patients infected with the outbreak strain.
Case Finding
We reviewed medical records of hospital A patients reported with CR E. coli isolates to identify epidemiologic links, including: diagnoses, inpatient days during the 6 months preceding positive culture, invasive medical procedures and surgeries, culture source, and antimicrobial susceptibility testing results.
We identified 842 AmpC E. coli isolates through retrospective surveillance since 2008 using clinical microbiology laboratory records, of which 63 (collected during February 2013–January 2014) were available for testing; laboratory data were not available for January–August 2010. Because we hypothesized that AmpC isolates might be related to the reported CR isolates, we selected 29 of the 63 AmpC E. coli isolates for PCR and PFGE testing on the basis of similarities to the reported CR E. coli patients, including: diagnosis of complicated biliary tract disease, hospital A inpatient stays during the 6 months preceding diagnosis, history of an invasive medical procedure or surgery, culture source, and resistance to fluoroquinolones and sulfonamides.
To estimate the prevalence of the outbreak strain among patients with complicated pancreatic or biliary disease, we cultured bile specimens obtained from hospitalized patients undergoing ERCP from December 1, 2013 through March 1, 2014.
To determine the extent of the outbreak strain among other acutely ill hospital A patients, we collected perianal swabs on all admissions to the critical care unit during January 1–March 3, 2014. This investigation was reviewed by the Centers for Disease Control and Prevention for human subjects’ protection and determined nonresearch.
Laboratory Testing
CR resistance was defined according to the 2013 Clinical and Laboratory Standards Institute breakpoints. Relatedness among CR and AmpC E. coli isolates was established using PFGE testing at the Washington State Public Health Laboratory. Isolates were considered indistinguishable if there was no band difference, closely related if a 2- or 3-band difference, possibly related if a 4- to 6-band difference, and unrelated if a band difference of 7 or greater.Reference Tenover, Arbeit and Goering 17 PCR screening for bla CMY and sequencing of bla CMY, fumC, and fimH amplicons were performed at Seattle Children’s Research Institute. Outbreak isolates featured a variant bla CMY-2 allele with a silent nucleotide substitution at position 660, fumC allele 41, and fimH allele 191.
We grouped PFGE results into 2 categories: unrelated (≥7-band difference) or related (indistinguishable, closely related, and possibly related) with differences of 0–2 independent genetic events.Reference Tenover, Arbeit and Goering 17 PFGE had a sensitivity of 90%, specificity of 100%, positive predictive value of 93%, and negative predictive value of 93% for presence of the bla CMY-2 variant. Isolates identified from CR surveillance and retrospective AmpC surveillance were tested using PFGE and PCR; because of the strong agreement between PFGE and PCR results, prospective surveillance and critical care unit prevalence isolates had PFGE testing only.
Endoscope Evaluation
The endoscope manufacturer was requested to assess reprocessing procedures and use of recommended cleaning and reprocessing techniques. Reprocessing (high-level disinfection after each endoscope’s use) includes manual cleaning, automated cleaning, and complete drying before the next use. The manufacturer observed ERCP technicians manually cleaning and inspecting the scopes. All 8 of hospital A’s ERCP scopes were sent to the manufacturer for evaluation of potential mechanical problems.
Hospital A’s automated endoscope reprocessor machine models DSD EDGE and CER series (both, Medivators) used in the final endoscope reprocessing step were also evaluated by the manufacturer. Data automatically recorded from all automated endoscope reprocessor machine alarms that occurred during June 2012–November 2013 were reviewed. Hospital A’s infection control practitioners interviewed endoscopy and clinical staff regarding hand hygiene and observed endoscope and procedure room cleaning.
Hospital A’s clinical microbiology laboratory cultured all 60 endoscopes after reprocessing on November 22, 2013, to identify bacterial contamination. Eight ERCP scopes were included in this sampling: 4 Olympus model V-Scope TJF-160VF and 4 Olympus TJF-Q180V (Olympus). We followed a modification of the endoscope testing protocol developed by the Centers for Disease Control and Prevention (unpublished), which included samples from the elevator, biopsy, and suction channels; port openings; water flushed through all ports; and the biopsy valve cap. Scopes having bacterial growth were reprocessed, recultured, and not used until results were negative. All ERCP scopes were cultured 1 time per week during the early phase of the investigation. Beginning January 22, 2014, ERCP scopes were cultured after every reprocessing and not used until culture results were negative for pathogenic bacteria at 48 hours.
Environmental Testing
Sink drains, faucets, and ports near the endoscopic reprocessing area, and basins and connectors within each of the 4 automated endoscope reprocessor machines in hospital A’s endoscopy suite 1, were cultured on November 22, 2013.Reference Palmore and Henderson 3 – Reference Lerner, Adler, Abu-Hanna, Meitus, Navon-Venezia and Carmeli 5
RESULTS
Case Finding and Case Description
Laboratory data indicated an increasing incidence of phenotypic AmpC E. coli infections at hospital A since 2008 (Figure 1). During August 2010–February 2014, 676 phenotypic AmpC E. coli isolates were identified from inpatients and outpatients. Of these, 479 (71%) were urine isolates.

FIGURE 1 AmpC phenotypic Escherichia coli isolates identified at hospital A, 2008–2013.
Forty-nine phenotypic AmpC E. coli isolates were obtained for testing: 10 CR isolates and 29 retrospectively collected isolates underwent PCR and PFGE testing; 10 prospective surveillance isolates underwent PFGE testing only.
Thirty-five (71%) of the 49 isolates met case definition (Figure 2), including all 10 CR E. coli (Figure 3). Three case isolates were removed from further analysis because they were from previously identified case patients, leaving 32 case patients. No surveillance isolates from critical care unit patients met case definition. Two patient isolates with the unique bla CMY-2, fumC, and fimH alleles identical to the outbreak profile had unrelated PFGE patterns (Figure 4). Case isolates fell into 2 related PFGE clusters. Each cluster contained isolates that were indistinguishable from each other, as well as other closely related isolates. Characteristics of case and non-case patients are shown in Table 1. All cases had complicated biliary or pancreatic disease and had undergone ERCP with biliary stent placement at hospital A since July 2010; certain patients also had previously undergone ERCP procedures at other hospitals. One bile isolate was obtained on the day of the patient’s first ERCP at hospital A, but all other patients had ERCP procedures performed at hospital A before isolation of phenotypic AmpC E. coli, with a median of 2 previous procedures (range, 1–12). Most patients were hospitalized at hospital A within the 6 months preceding their infection diagnosis.
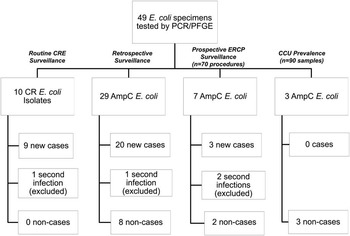
FIGURE 2 Schematic of all tested AmpC and carbapenem-resistant (CR) Escherichia coli isolates from hospital A by route of isolate identification and case status. CCU, critical care unit; CRE, carbapenem-resistant E. coli; ERCP, endoscopic retrograde cholangiopancreatography; PCR, polymerase chain reaction; PFGE, pulsed-field gel electrophoresis.
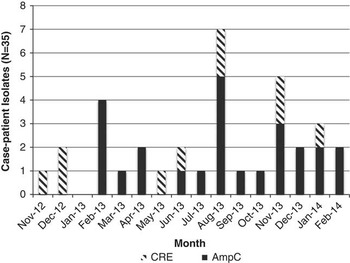
FIGURE 3 All AmpC Escherichia coli case-isolates identified at hospital A, November 2012–February 2014. CR, carbapenem-resistant.
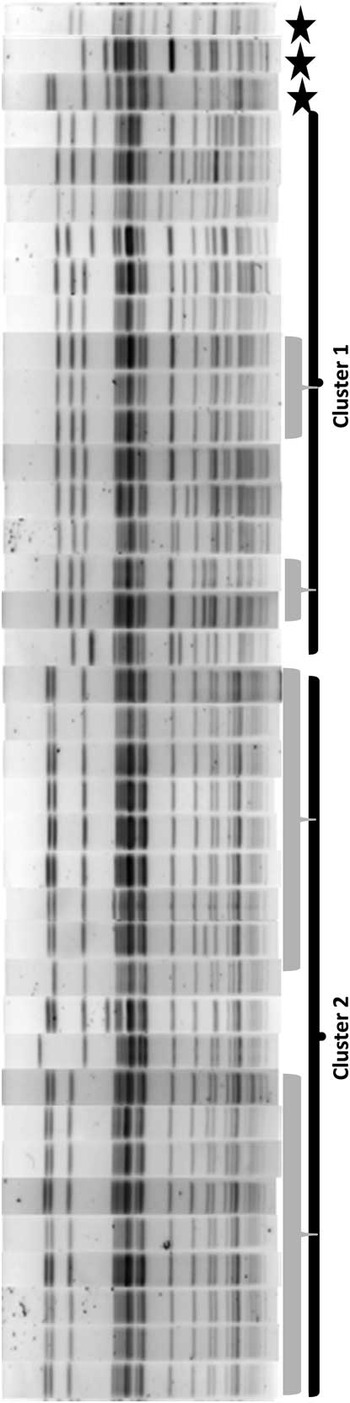
FIGURE 4 Pulsed-field gel electrophoresis results for 35 case-patient isolates and 3 endoscope isolates obtained November 2012–April 2014 from hospital A, Washington State. Isolates within each gray bar are defined as indistinguishable from each other (0 band differences); isolates within each cluster bar (cluster 1 and cluster 2) are defined as closely (2–3 band differences) or possibly (4–6 band differences) related to each other; and cluster 1 is related to cluster 2. Stars distinguish isolates identified as cases through polymerase chain reaction analysis that were not related on pulsed-field gel electrophoresis.
TABLE 1 Characteristics of AmpC Escherichia coli Case Patients vs Non-case Patients, Hospital A, Washington State
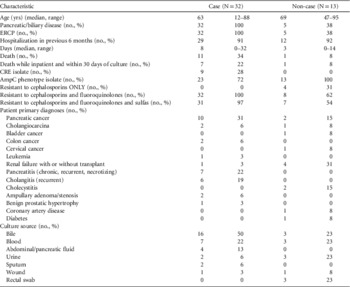
NOTE. CRE, carbapenem-resistant Enterobacteriaceae; ERCP, endoscopic retrograde cholangiopancreatography.
Of 32 case patients, 11 (34%) died during the investigation (Figure 5); 7 (64%) of the deaths occurred during hospitalization within 30 days of the date that the E. coli isolate of interest was obtained (median, 5 days between isolate collection and death [range, 2–24 days]). Of these 7 deaths, 5 occurred among the 9 patients with CR E. coli (56% mortality) and 2 occurred among the 23 patients with AmpC E. coli (9% mortality); the difference in mortality rates between patients with CR versus non-CR AmpC E. coli infection was significant (P=.004). The primary diagnoses for the 7 patients who died included pancreatic cancer (3), colon cancer (2), primary sclerosing cholangitis (1), and renal/pancreatic transplant (1).

FIGURE 5 Mortality among 32 case-patients. Acute deaths occurred during same hospitalization and within 30 days of isolate collection. CR, carbapenem-resistant.
Of the 49 patient isolates, 14 (29%) did not meet the case definition (Figure 2), including all the AmpC prospective prevalence study isolates. One non-case patient isolate was eliminated from further analysis because it was a second culture from an already identified patient; therefore there were 13 total non-case patients. Non-case patient characteristics are shown in Table 1. Of the 13 non-case patients, 1 (8%) died in hospital 10 days after the phenotypic AmpC E. coli isolate was obtained, with a diagnosis of chronic renal failure complicated by congestive heart failure.
Endoscope Evaluation
The endoscope manufacturer’s review determined that endoscope reprocessing procedures at hospital A were above the industry standard, and all technicians performed manual endoscope cleaning in a manner consistent with manufacturer guidelines. Evaluation of the automated endoscope reprocessors revealed no defects; all were operating correctly. Seven of the 8 ERCP scopes submitted to the manufacturer had at least 1 critical defect requiring repair that had not been detected by the facility (Table 2), including 3 scopes that had passed the leak test at hospital A but failed at the manufacturer.
TABLE 2 Pathogenic Bacteria Isolated From Endoscopic Retrograde Cholangiopancreatography (ERCP)–Associated Scopes, Hospital A, Washington State

NOTE. This table includes all ERCP-associated scopes with substantial bacterial contamination after reprocessing, including the 8 original ERCP scopes and the additional ERCP scopes purchased during the investigation. Each scope listed in the first column is a different scope. Scopes are used repeatedly and cultured after each use; therefore, 1 scope might have been contaminated multiple times. E. coli, Escherichia coli; MDR, multidrug-resistant; MSSA, methicillin-sensitive Staphylococcus aureus; PFGE, pulsed-field gel electrophoresis.
Among the 60 scopes cultured on November 22, 2013, a total of 4 (7%) had gram-negative bacteria isolated, including 2 ERCP scopes that had AmpC E. coli contamination (Table 2) as well as 2 colonoscopes growing Pseudomonas and Roseomonas species. The 2 ERCP scopes harbored 4 different strains of AmpC E. coli cultured from different locations on the scopes; 2 of the isolates were related to patient isolates by PFGE and exhibited the outbreak gene sequence profile.
Different technicians had cleaned the contaminated scopes in the same endoscope-reprocessing area. Cultures from environmental samples obtained in and around the scope cleaning area were negative for bacterial growth.
During routine ERCP scope culturing from January 22–May 14, 2014, a total of 365 cultures were collected, of which 65 (18%) were positive for bacterial growth. Of these 65 cultures with bacterial growth, 55 (85%) grew common skin flora considered contaminants and 10 (15%) grew 11 pathogenic bacteria from swab samples taken from the scopes’ elevator channels (Table 2).
DISCUSSION
We describe an outbreak of non–carbapenemase-producing CR and AmpC E. coli infections associated with ERCP procedures among patients with complicated pancreatic and biliary disease. Especially disturbing is that the outbreak occurred despite no identified breaches in reprocessing of the endoscopes. Even after enhancing endoscope reprocessing with meticulous manual cleaning, enteric bacteria continued to be recovered from endoscope elevator channels. In addition, endoscopes without apparent defects or indications for servicing were identified as having critical abnormalities when submitted to the manufacturer for evaluation. Although the majority of the scopes required such servicing, both those needing and those not needing repairs harbored pathogens from the elevator channel, and positive cultures were obtained from scopes even after overhaul by the manufacturer. Occult mechanical defects, as well as inherent difficulty in cleaning and decontaminating the endoscope elevator channel, may have facilitated transmission of the outbreak strain. No routine endoscope maintenance or servicing guidelines are available from the manufacturers. Our results indicate that routine endoscope evaluation and maintenance schedules might need to be included in the approval process for these devices.
Our investigation identified no evidence of a reservoir of the outbreak strain either among other critically ill patients in the unit to which such patients were routinely admitted or in the environment; only 2 of our AmpC (non-CR) E. coli patients had a history of previous admission to long-term-care facilities.
ERCP-associated outbreaks have been previously reported, including those caused by Pseudomonas aeruginosa, Reference Struelens, Rost and Deplano 18 – Reference Alfa, Olson, DeGagne and Jackson 27 MDR Klebsiella pneumoniae, Reference Aumeran, Poincloux and Souweine 26 and more recently, CR Enterobacteriaceae.Reference Gastmeier and Vonberg 12 , Reference Epstein, Hunter and Arwady 16 Previous reports of ERCP endoscope–related outbreaks have been linked to inadequate endoscope cleaning. For example, in 1 report the contaminated portion of an ERCP scope was a blind channel that was not cleaned during the disinfection phase; the problem was alleviated by cleansing and disinfecting this channel and adding an isopropanol-air flush of all channels.Reference Struelens, Rost and Deplano 18 A 2002 survey of reprocessing methods reported that only 43% of healthcare centers were following all guidelines for scope reprocessing.Reference Alfa, Olson, DeGagne and Jackson 27 In particular, the mobile elevator channel can be difficult to clean; 1 study reported that 19% of elevator channels were inadequately cleaned.Reference Alfa, Olson, DeGagne and Simner 28 In contrast, in both our outbreak and a recent CR E. coli outbreak,Reference Epstein, Hunter and Arwady 16 transmission of a unique organism was linked to an ERCP scope despite scope cleaning and disinfecting processes that followed manufacturer guidelines and/or exceeded industry standards.
By definition, case isolates recovered during this outbreak revealed a novel bla CMY-2 allele in a distinctive fumC/fimH strain background. The patient isolates exhibited resistance not only to third-generation cephalosporins but also to other classes of antibiotics (fluoroquinolones, sulfonamides, aminoglycosides), suggesting that unlike chromosomal AmpC producers (eg, Enterobacter species), the AmpC determinant was likely borne on a multidrug-resistant plasmid. We hypothesize that the carbapenem-sensitive AmpC E. coli isolates then gave rise to CR E. coli isolates by way of porin defects occurring under strong selection pressure, most likely due to antibiotic exposure.
This outbreak highlights the importance of public health surveillance for identifying MDR outbreaks, and the useful addition of molecular methods for identifying resistance mechanisms and characterizing strains. Considerable attention has been paid to detection and prevention of carbapenemase-producing CR Enterobacteriaceae Reference Gupta, Limbago, Patel and Kallen 29 , Reference Kallen and Guh 30 because of the severity of these infections and limited treatment options.Reference Van Duin, Kaye, Neuner and Bonomo 31 Although outbreaks associated with non–carbapenemase-producing CR Enterobacteriaceae have been uncommonly reported,Reference Ktari, Arlet and Verdet 32 – Reference Arena, Giani and Becucci 35 our investigation indicates that the clinical significance might be similar to that of carbapenemase-producing strains. Additionally, the outbreak was detected through a public health surveillance program that was enhanced with the addition of molecular testing and would likely have gone undetected otherwise. Routine surveillance is crucial for promptly recognizing outbreaks and monitoring and responding to the ongoing threat from MDR organisms in healthcare facilities.Reference Kallen and Guh 30 , Reference Fitzpatrick, Zembower, Malczynski, Qi and Bolon 36
Because public health agencies cannot monitor all MDR organisms, healthcare facilities should consider implementing internal surveillance for clinically significant organisms (and clusters of infections) even when reporting to public health agencies is not required. Reporting of specific MDR organisms to public health authorities should be based on the organism’s public health significance, the local epidemiology, and the need for public health action.
As a result of this outbreak, hospital A undertook costly and extraordinary measures to minimize risk for endoscope-related infection transmission. The facility now quarantines ERCP scopes after cleaning and reprocessing and does not release them for use until cultures from the elevator channel are negative at 48 hours after culture plating. Despite ongoing careful manual and automated cleaning at hospital A, 3% of scopes continue to have positive pathogenic cultures and require additional cleaning before the next use.
Our report has limitations. Our investigation was initiated after certain preventive measures had been implemented by hospital A; therefore, we were unable to observe practices at the time transmissions occurred. Cases were identified retrospectively on the basis of having severe pancreatic or biliary disease, as well as the availability of microbiology records and an isolate for molecular laboratory testing; therefore, our investigation might not represent all persons who carried the outbreak strain. Additionally, our prospective surveillance focused on patients at high risk. Because affected patients all had severe underlying disease, the role of the outbreak strain in patient deaths cannot be determined.
Further study of the adequacy of endoscope reprocessing is needed, particularly for scopes with elevator channels. On the basis of our experience, we suspect endoscope-associated transmission of pathogenic bacteria might be both more common than recognized and not adequately prevented by current endoscope reprocessing guidelines. Public health authorities, regulatory agencies, and endoscope manufacturers should consider evaluating the adequacy of reprocessing standards and endoscope design to identify improved strategies for endoscope decontamination. Endoscopists should inform patients of the risk for endoscope-associated transmission of MDR organism infection along with other known endoscopy-associated risks when obtaining consent for procedures.
ACKNOWLEDGMENTS
We thank the Washington State Department of Health and the Centers for Disease Control and Prevention.
Financial support. Investigative work was performed by Public Health–Seattle and King County and PFGE was performed by Washington State Department of Health. Virginia Mason Medical Center paid for the laboratory testing and materials used in the hospital during the investigation.
Potential conflicts of interest. All authors report no conflicts of interest relevant to this article.
Disclaimer. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.









