Introduction
Bacidia De Not. is an almost exclusively epiphytic crustose lichen genus, characterized by smooth or warted to granular thalli; acicular multiseptate spores with an average of 30–40, up to 100 μm in length and 2.0–4.0 μm in width; and a pronounced exciple, consisting of radiating hyphae, frequently with enlarged lumina cells at the tips (Ekman Reference Ekman1996; Llop Reference Llop2007). Until recently, a large variety of lichens were included in the genus Bacidia in a broad sense. Zahlbruckner (Reference Zahlbruckner1921–1940) used the name for crustose lichens with a chlorococcoid photobiont, biatorine apothecia, and ascospores with three or more transverse septa. This circumscription was unnatural and included numerous taxa not closely related to the type species, Bacidia rosella (Pers.) De Not. (Gerasimova & Ekman Reference Gerasimova and Ekman2017).
The first extensive phylogenetic study of Bacidia was based on gene sequences of the nuclear ribosomal internal transcribed spacer (nrITS). It resulted in a re-evaluation of the generic delimitation, suggesting the transfer of several species to genera such as Biatora Fr., Toninia A. Massal. and Bacidina Vězda (Ekman Reference Ekman2001). In more recent phylogenetic studies, several species have been transferred to further genera, such as Bellicidia Kistenich et al., Bibbya J. H. Willis, Scutula Tul. and Toniniopsis Frey (Kistenich et al. Reference Kistenich, Timdal, Bendiksby and Ekman2018). These phylogenetic studies narrowed the generic concept, although the relationship between species, and support for the species groups, remained unclear. Several additional studies, partially focusing on Bacidia, used two or multilocus data sets, combining sequences from nrITS with mitochondrial small subunit rDNA (mtSSU) (Andersen & Ekman Reference Andersen and Ekman2005; Lendemer et al. Reference Lendemer, Harris and Ladd2016; Malíček et al. Reference Malíček, Palice, Vondrák, Łubek and Kukwa2018), and nuclear ribosomal large subunit rDNA (nrLSU) with mtSSU (Lumbsch et al. Reference Lumbsch, Schmitt, Palice, Wiklund, Ekman and Wedin2004; Sérusiaux et al. Reference Sérusiaux, van den Boom, Brand, Coppins and Magain2012), but they covered only a small number of species of Bacidia s. str. Additional multilocus phylogenetic studies involving two genes encoding the largest and the second-largest subunit of RNA polymerase II (RPB1 and RPB2, respectively) also encompassed only six species, namely B. absistens (Nyl.) Arnold, B. arceutina (Ach.) Arnold, B. squamulosula (Nyl.) Zahlbr., B. schweinitzii (Fr. ex Tuck.) A. Schneid., B. rosella and B. rubella (Hoffm.) A. Massal. (James et al. Reference James, Kauff, Schoch, Matheny, Hofstetter, Cox, Celio, Gueidan, Fraker and Miadlikowska2006; Miadlikowska et al. Reference Miadlikowska, Kauff, Hofstetter, Fraker, Grube, Hafellner, Reeb, Hodkinson, Kukwa and Lücking2006, Reference Miadlikowska, Kauff, Högnabba, Oliver, Molnár, Fraker, Gaya, Hafellner, Hofstetter and Gueidan2014; Reese Næsborg et al. Reference Reese Næsborg, Ekman and Tibell2007; Ekman et al. Reference Ekman, Andersen and Wedin2008; Kistenich et al. Reference Kistenich, Timdal, Bendiksby and Ekman2018).
In the first molecular study on Bacidia s. str. from the Russian Far East (RFE), taxon sampling was expanded (Gerasimova et al. Reference Gerasimova, Ezhkin and Beck2018). Four new species were described, and these are currently considered endemic. While well-supported groups within Bacidia s. str. were revealed, support in the nodes of basal clades was low and several polytomies remained.
The main goal of this study was to investigate whether a phylogeny of Bacidia s. str. inferred from a larger, multilocus data set with a broader taxon sampling would yield higher backbone support than the previous studies based on nrITS sequences. We address the phylogeny of Bacidia s. str. based on nrITS data together with newly obtained data from nrLSU, mtSSU and two protein-coding genes, RPB1 and RPB2, and 131 sequences of Bacidia currently available in GenBank.
Material and Methods
Specimens
This study was based on fresh collections of Bacidia gathered by the authors in the field from 2013 to 2015 in the southern part of the Russian Far East (Primorskiy and Khabarovskiy Krai), and on Sakhalin and the Kurile Islands (as indicated in Gerasimova et al. (Reference Gerasimova, Ezhkin and Beck2018)). The second author collected the Bacidia obtecta specimens on Sakhalin in 2017 (see details under Taxonomy). Voucher specimens are deposited in the herbaria of the Botanische Staatssammlung München (M), Institute of Marine Geology and Geophysics (SAK) and Altai State University (ALTB). The inclusion of Bacidia squamulosula was based on a specimen examined in M (M-0012326; K. Kalb, Lichenes Neotropici No. 405, Bacidiopsora squamulosula (Nyl.) Kalb). Detailed information on the newly obtained sequences, together with their respective voucher information and GenBank Accession numbers, is given in Table 1.
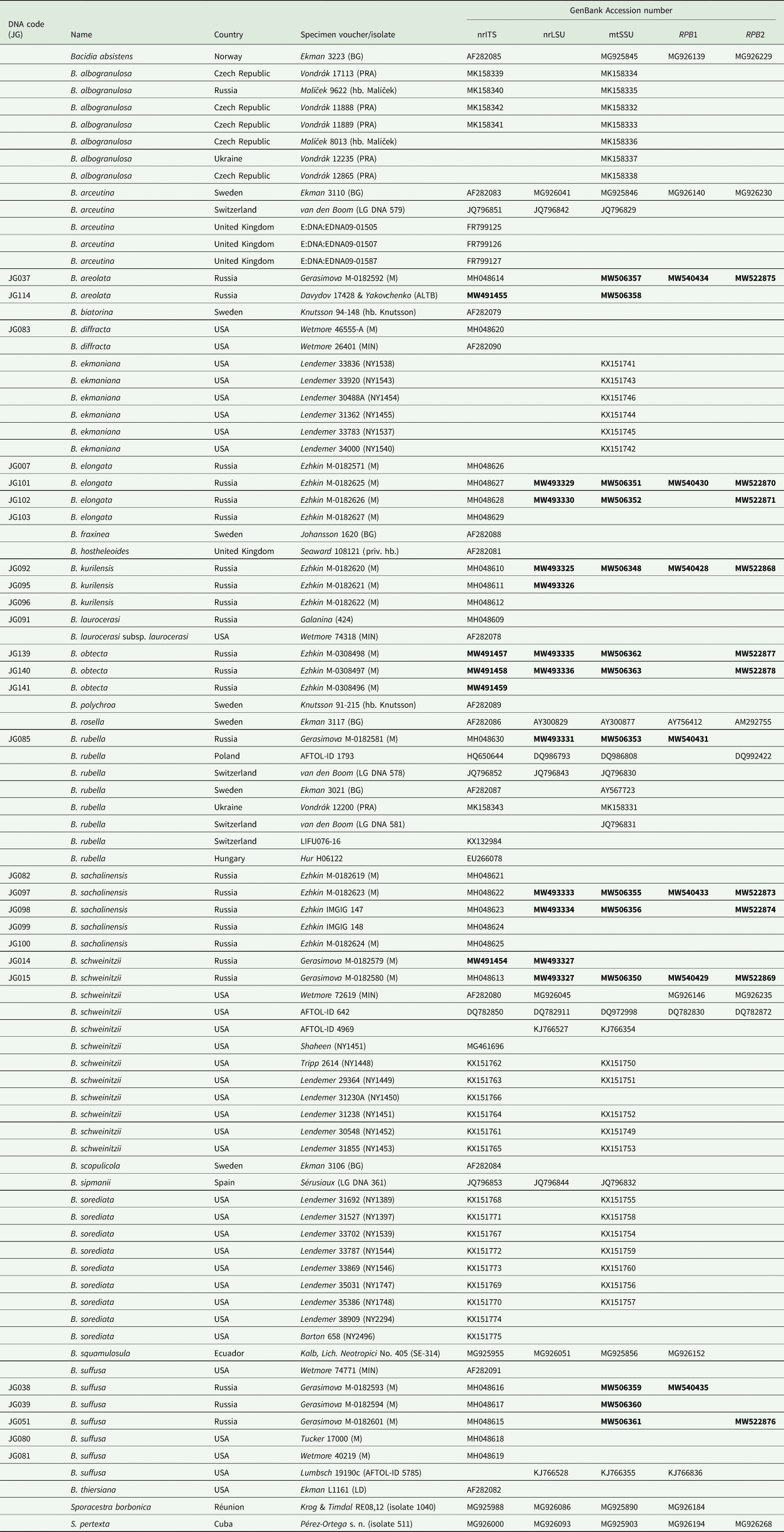
Table 1. DNA codes and specimen information used in this study for species of Bacidia and outgroup taxa, with their respective GenBank Accession numbers. New sequences are in bold.
Morphology
Microscopic observations were made using a Zeiss Axioplan light microscope (Oberkochen, Germany) equipped with differential interference contrast. Cross-sections of apothecia were made on a Leica Jung Histoslide 2000 Mikrotom (Heidelberg, Germany), with a thickness of 8–10 μm. Micrographs of cross-sections were taken on a Zeiss Axioplan with an attached AxioCam 512 Color camera and processed with the Zeiss ZEN 2.3 (blue edition) image software. Macrographs of external characters were taken on a Leica Z6 Apo microscope (with a ×1.0 Planapo lens; Leica, Germany) with a Sony Alpha 6400 camera (Sony, Japan) attached and equipped with a StackShot Macro Rail (Cognisys, USA) as detailed in Gerasimova et al. (Reference Gerasimova, Urbanavichene, Urbanavichus and Beck2021). A single image was mounted from 40–100 serial images using Helicon Focus v.7 software (Helicon, USA).
Measurements are given as (min–) average ± SD (–max) (SD = standard deviation, n 1 = number of all observations, n 2 = number of specimens observed). We provide a detailed description of specimens using standard microscopic techniques following Ekman (Reference Ekman1996) and use the proper exciple subdivision scheme, differentiating the following structures: the rim, lateral part and medullary part. Pigment characterization follows Meyer & Printzen (Reference Meyer and Printzen2000) and Ekman (Reference Ekman1996).
DNA extraction, PCR amplification and DNA sequencing
DNA extraction was carried out using the Stratec Invisorb Spin Plant Mini Kit (Stratec Molecular GmbH, Berlin, Germany) following the manufacturer's instructions. Five to eight apothecia were used from fresh material not older than five years. Thallus fragments were removed to minimize the risk of contamination by, for example, lichenicolous fungi. PCR amplification, purification and sequencing were performed as described in Gerasimova et al. (Reference Gerasimova, Ezhkin and Beck2018). Cycling conditions included initial denaturation at 95 °C for 2 min, 5 cycles of 95 °C for 40 s, 54 °C for 60 s, 72 °C for 90 s, 33 cycles of 95 °C for 40 s, 54 °C for 60 s, 72 °C for 90 s, and a final extension step at 72 °C for 7 min. In those cases when PCR products were insufficient, a second PCR with a reduced number of cycles was conducted: denaturation at 95 °C for 2 min, 5 cycles of 95 °C for 40 s, 54 °C for 60 s and 72 °C for 90 s, 22 cycles of 95 °C for 40 s, 54 °C for 60 s and 72 °C for 90 s, with a final extension step at 72 °C for 7 min. We used five pairs of primers: ITS1F (White et al. Reference White, Bruns, Lee, Taylor, Innis, Gelfand, Sninsky and White1990) and ITS4m (Beck & Mayr Reference Beck and Mayr2012), LR0R (Rehner & Samuels Reference Rehner and Samuels1994) and LR5 (Vilgalys & Hester Reference Vilgalys and Hester1990), mtSSU1 and mtSSU3R (Zoller et al. Reference Zoller, Scheidegger and Sperisen1999), fRPB2-5F and fRPB2-7cR (Liu et al. Reference Liu, Whelen and Hall1999), and newly designed primers for Bacidia gRPB1AFba (5′-GAG TGY CCG GGA CAT TTT GG-3′) and fRPB1cRba2 (5′-GSC CRG CAA TRT CGT TAT CCA-3′).
Alignment and phylogenetic analyses
Forty-eight new sequences of Bacidia s. str. from three ribosomal RNA-coding (nrITS, nrLSU, mtSSU) and two protein-coding genes (RPB1 and RPB2) were obtained and combined with 22 nrITS sequences from our previous study and 109 sequences of Bacidia s. str. from GenBank (Table 1). Bacidia schweinitzii (JG014), B. areolata Gerasimova & A. Beck (JG114) and B. obtecta sp. nov. (JG139-141) were newly sequenced.
Altogether 85 individuals from 24 OTUs of Bacidia s. str. were included. ‘Phyllopsora’ borbonica Timdal & Krog and Sporacestra pertexta (Nyl.) Stapnes & Timdal were selected as the outgroup based on the results of Kistenich et al. (Reference Kistenich, Timdal, Bendiksby and Ekman2018).
BLAST searches in GenBank were performed to detect and exclude accessory/lichenicolous fungi and potential contaminations. Alignments were carried out using standard settings in MUSCLE v.3.8.31 (Edgar Reference Edgar2004) as implemented in PhyDE-1 v.0.9971 and optimized manually. Positions where a gap had to be inserted in more than 95% of the sequences were excluded. The most variable ITS1 region was aligned using Gblocks v.0.91.1 (https://ngphylogeny.fr/tools/tool/276/form). The alignments are provided in Supplementary Material File S1 (available online).
Sequences of Bacidia friesiana (Hepp) Körb., B. purpurans R. C. Harris et al., B. hostheleoides (Nyl.) Zahlbr. and B. thiersiana Lendemer (deposited as Bacidia lutescens Malme in GenBank) were revealed as separate lineages. Bacidia purpurans is more distantly related to Bacidia s. str. than the outgroup (Supplementary Material File S2, Figs S1–S3, available online). According to a BLAST search, B. friesiana (MH539765) is closest to Bacidina with more than 90% similarity. Given the high morphological variation and the necessity for further sequencing, we refrain from transferring this species to another genus.
We performed analyses for single-locus and concatenated data sets as follows: 1) single-locus data set for each locus separately (trees are included in Supplementary Material File S3, Figs S5–S19, available online); 2) concatenated multilocus data set including 26 taxa (including the outgroup) with sequences from three to all five genes available for each specimen (both newly obtained and GenBank sequences (Fig. 1; Supplementary Material File S3, Figs S1–S2, available online); 3) concatenated all-taxa data set including 87 taxa (including the outgroup) with a minimum of one sequence available (both newly obtained and GenBank sequences) in order to include all specimens with sequences available (Fig. 2; Supplementary Material File S3, Figs S3–S4, available online).
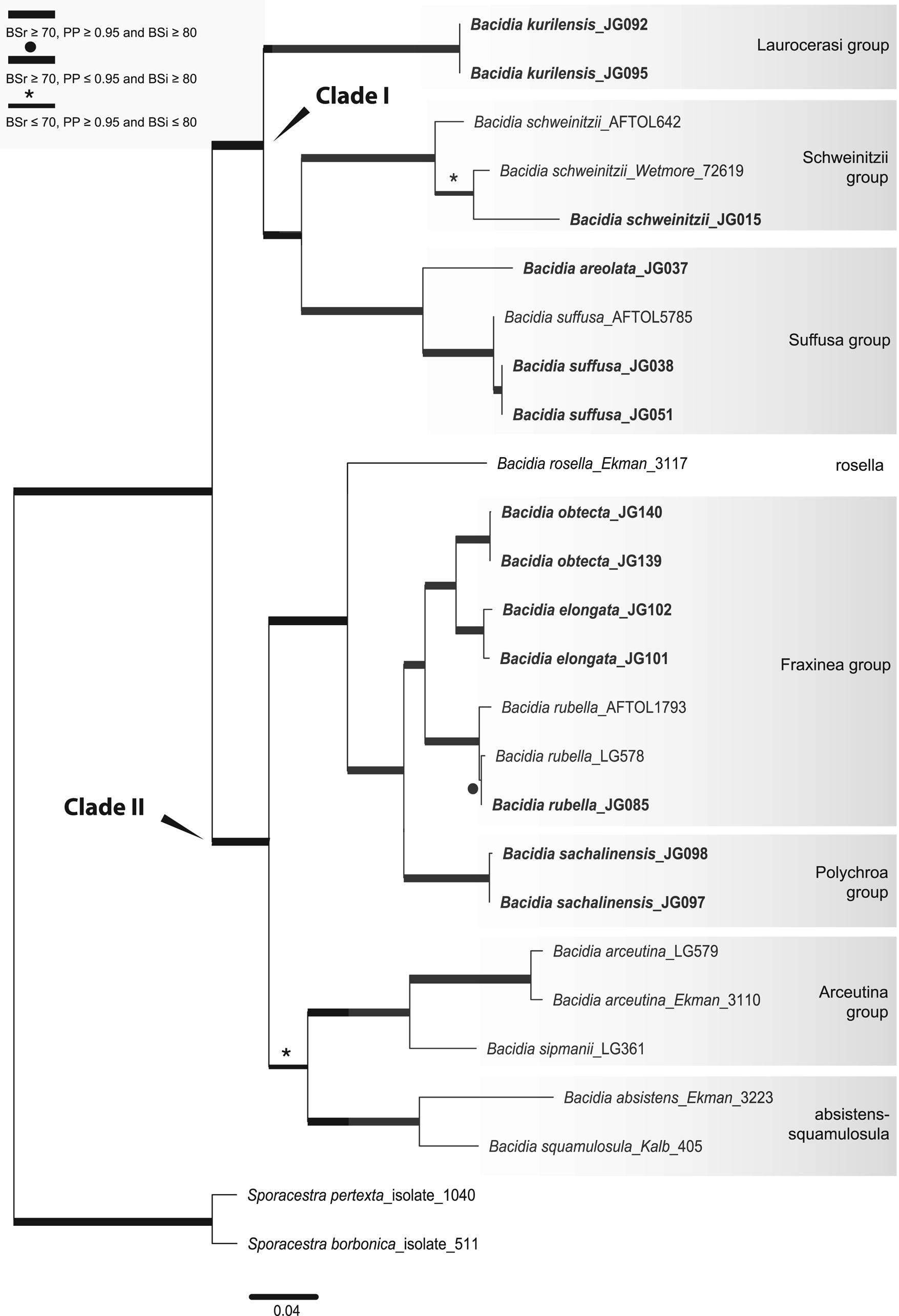
Fig. 1. Maximum likelihood (ML) tree of Bacidia s. str. resulting from the RAxML analysis of the concatenated multilocus data set with a minimum of three loci included (out of nrITS, nrLSU, mtSSU, RPB1 and RPB2). RAxML bootstrap values (BSr), Bayesian posterior probabilities (PP) and IQ-TREE bootstrap values (BSi) are indicated. Highly supported branches with BSr ≥ 70%, PP ≥ 0.95 and BSi ≥ 80% are marked in bold; strongly supported branches with BSr ≥ 70% and BSi ≥ 80% are also marked in bold with a black dot above the branch; branches with PP ≥ 0.95 are marked in narrower bold lines and a star above the branches. Sporacestra taxa are the outgroup. Bacidia lutescens is referred to as B. thiersiana in the text.
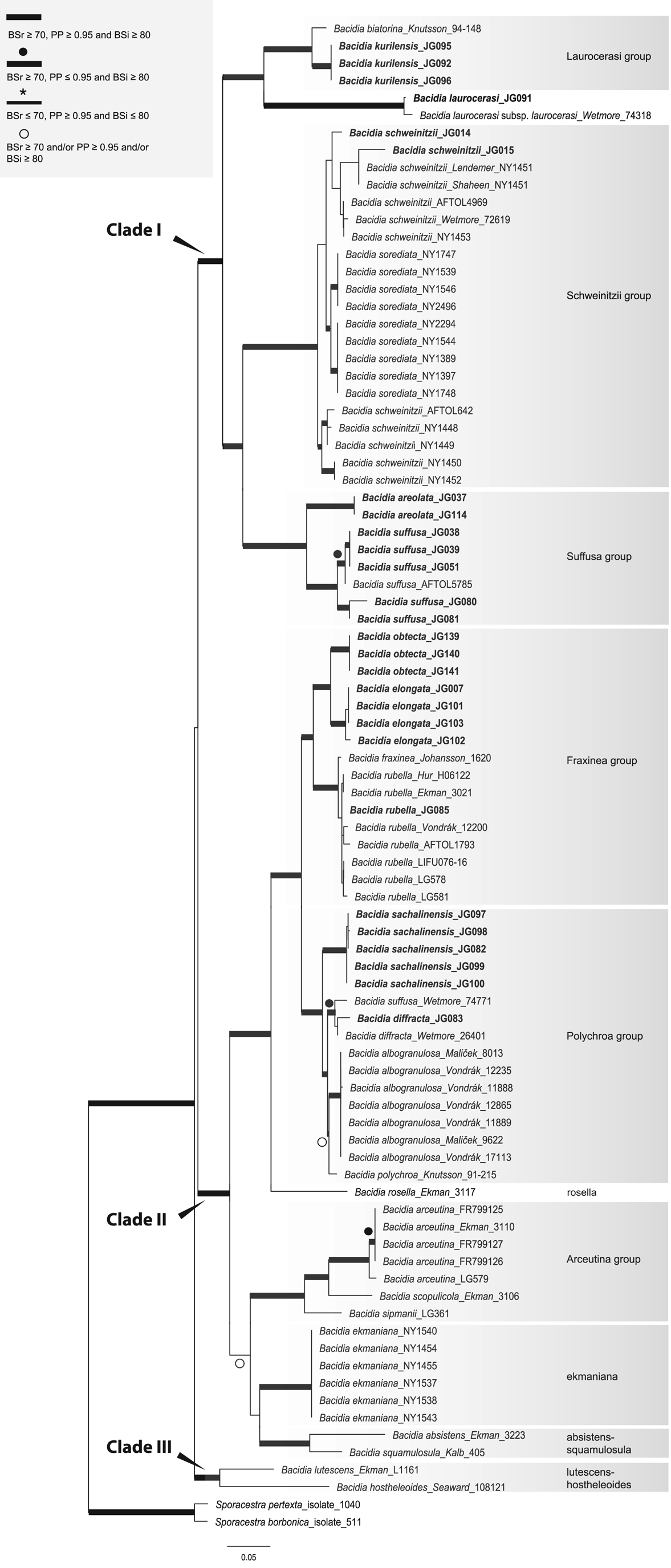
Fig. 2. Maximum likelihood (ML) tree of Bacidia s. str. resulting from the RAxML analysis of the concatenated all-taxa data set with a minimum of one sequence available (out of nrITS, nrLSU, mtSSU, RPB1 and RPB2). RAxML bootstrap values (BSr), Bayesian posterior probabilities (PP) and IQ-TREE bootstrap values (BSi) are indicated. Highly supported branches with BSr ≥ 70%, PP ≥ 0.95, and BSi ≥ 80% are marked in bold; strongly supported branches with BSr ≥ 70% and BSi ≥ 80% are also marked in bold with a black dot above the branch; branches with PP ≥ 0.95 are marked in narrower bold lines and a star above the branches; branches with PP ≥ 0.95, and/or BSr ≥ 70% and/or BSi ≥ 80% are marked with a white dot. Sporacestra taxa are the outgroup. Bacidia lutescens is referred to as B. thiersiana in the text.
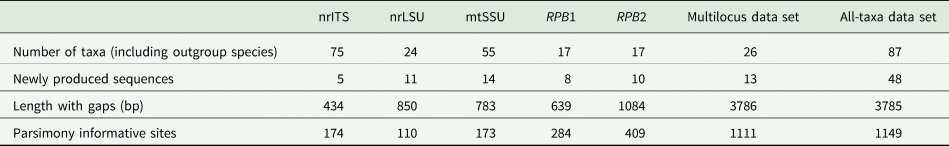
Based on the support values of the phylogenetic analyses, no supported incongruence between the single-locus tree topologies was found; therefore, the concatenated alignment was analyzed. The number of taxa and alignment details for the single and concatenated data sets are summarized in Table 2. We used the multilocus alignment to produce a robust phylogeny, including the taxa with sequences from a minimum of three available loci. For comparison, we analyzed the all-taxa data set with all Bacidia s. str. specimens, even if only the sequence of a single locus was available.
Table 2. Overview of the numbers of taxa and newly produced sequences for each genetic marker and concatenated alignment for Bacidia in this study.
The alignment with nrITS, nrLSU, mtSSU, RPB1 and RPB2 was subjected to Bayesian inference (BI) and maximum likelihood (RAxML and IQ-TREE) analyses for the single and concatenated data sets separately, as implemented in Gerasimova et al. (Reference Gerasimova, Urbanavichene, Urbanavichus and Beck2021).
Substitution models for the entire (no-partitioned) concatenated and single-locus data sets were selected using jModelTest v.2 (Darriba et al. Reference Darriba, Taboada, Doallo and Posada2012) following the Akaike selection criterion (AIC). The best substitution model was selected in jModelTest with 1-, 2- and 6-model groups with additional options of independent state (nucleotide) frequencies, gamma-distributed rate variations across sites and a proportion of invariable sites (24 possible models).
Bayesian inference was carried out in MrBayes v.3.2.6 (Ronquist et al. Reference Ronquist, Teslenko, van der Mark, Ayres, Darling, Höhna, Larget, Liu, Suchard and Ronquist2012) using the Markov chain Monte Carlo method (MCMC) and GTR + I + G model. Two parallel runs were performed (two cold chains) with a single tree saved every 10th generation for a total of 1 000 000 generations. According to the trends in likelihood values, the convergence of the Markov chain was reached after 10 000 generations. As a result, the initial 10% was discarded as burn-in and the results summarized as a 50% majority-rule consensus tree.
Maximum likelihood (ML) analysis was performed with RAxML v.8.2.4 on the CIPRES web portal (Miller et al. Reference Miller, Pfeiffer and Schwartz2010). The GTRGAMMA model with rapid bootstrap analysis searching for the best-scoring ML tree, using a majority-rule consensus tree and 1000 bootstrap iterations, was used for the entire data. The bipartition of the best-scoring tree was drawn onto the most likely tree topology following the recommendation of Stamatakis (Reference Stamatakis2014).
Further tree reconstruction using ML analysis was performed in IQ-TREE v.1.6.12 using standard bootstrap approximation with 1000 bootstraps, specifying the GTR + I + G model as suggested by jModelTest (Nguyen et al. Reference Nguyen, Schmidt, von Haeseler and Minh2015).
The phylogenetic trees were visualized using FigTree v.1.4.2 (Rambaut Reference Rambaut2009). The ML trees based on the different substitution models from single and concatenated data sets were congruent, therefore only the RAxML trees for the concatenated data sets are shown. Clades that received bootstrap support values (BSr) ≥ 70% in RAxML, posterior probabilities (PP) ≥ 0.95 in BI and bootstrap values (BSi) ≥ 80% in IQ-TREE were considered highly supported. The concatenated and individual gene trees obtained from RAxML, MrBayes and IQ-TREE are provided as Supplementary Material File S3 (Figs S1–S19).
Results
The BI and ML analyses for the single and concatenated data sets recovered highly concordant topologies of the phylogenetic trees. Both multilocus and all-taxa trees resulted in well-supported basal nodes (Figs 1 & 2).
Single-locus trees were congruent with the multilocus and all-taxa trees, but with high support in the terminal groups and low support in the backbone. All clades in the multilocus tree received strong support (i.e. with BSr ≥ 70%, PP ≥ 0.95 and BSi ≥ 80%) both in the terminal and deeper branches with only a small number of exceptions. The clade including B. arceutina, B. absistens and B. squamulosula was supported in BI (BSr/PP/BSi: 66/0.97/62); the subclade with two B. rubella from the RFE (JG085) and Switzerland (LG578) was supported in RAxML (BSr/PP/BSi: 87/0.75/71), and the subclade with B. schweinitzii from the RFE (JG015) and the USA (Wetmore 72619) was supported in BI only (BSr/PP/BSi: 56/0.99/61). The multilocus tree is congruent with the all-taxa phylogeny, forming the same groups with high support values (Fig. 2). The enlarged all-taxa phylogeny comprises all specimens with sequence data available; therefore, we discuss the groups in more detail based on this tree. According to phylogenetic results and morphological characters, six Bacidia s. str. groups are recognized: Laurocerasi, Schweinitzii, Suffusa, Fraxinea, Polychroa, and Arceutina. The separate lineages of 1) B. ekmaniana R. C. Harris et al., B. absistens and B. squamulosula, and 2) B. hostheleoides and B. thiersiana, are discussed individually. Furthermore, a correlation between these phylogenetic groups and the apothecial pigments was observed (see Table 3 and Discussion).
Table 3. Comparison of the pigmentation in the different phylogenetic groups in Bacidia s. str. as shown in Fig. 2. Pigment characterization follows Meyer & Printzen (Reference Meyer and Printzen2000) and Ekman (Reference Ekman1996). The three parts of the table represent the pigmentation of the two major clades and separate lineages of Bacidia s. str.
The Laurocerasi group, consisting of the three lineages B. biatorina (Körb.) Vain., B. kurilensis Gerasimova et al. and B. laurocerasi (Delise ex Duby) Zahlbr., received strong support (BSr/PP/BSi: 92/1.0/93).
Specimens of B. schweinitzii and B. sorediata Lendemer & R. C. Harris formed a paraphyletic clade (Schweinitzii group), comprising several separate clades with the highest support (BSr/PP/BSi: 99/1.0/100). Two subclades were present in B. schweinitzii, each containing an additional phylogenetic structure. One subclade, with high support (BSr/PP/BSi: 84/0.95/83), includes only GenBank sequences of B. schweinitzii from North America (AFTOL642 and NY). The second subclade contains both individuals from the RFE and North America and two subclades of B. sorediata, but without significant support (BSr/PP/BSi: 40/0.8/46). Individuals of B. schweinitzii from the RFE (JG014 and JG015) were found in two groups in the ITS phylogeny (see Supplementary Material File S3, Figs S5–S7, available online). Given the differences in nrITS sequences (3% = 13 nucleotides), they probably belong to two independent species (see Discussion below). This is also supported by morphological observations which indicate differences in thallus and apothecial characters (details are summarized in Supplementary Material File S4, available online). The Schweinitzii group is sister to the Suffusa group; together, they form a well-supported clade (BSr/PP/BSi: 78/1.0/79).
The clade containing B. areolata and B. suffusa (Fr.) A. Schneid. (Suffusa group) was highly supported (BSr/PP/BSi: 100/1.0/99). Two separate well-supported lineages in B. suffusa represent the North American and RFE populations. For two individuals from the USA, JG080 and JG081, only the nrITS2 sequences were available, most likely due to the age of the specimens (Gerasimova et al. Reference Gerasimova, Ezhkin and Beck2018). However, compared to the RFE population, they already reveal variations in the relatively conserved nrITS2 region: four transitions at several positions, indicating a 2% difference. For another individual from the USA (AFTOL5785), the nrITS sequence is unavailable, but there are nrLSU, mtSSU and RPB1 sequences. In the mtSSU phylogeny, it also forms a separate lineage from the RFE individual, JG051, with a 3% (= three nucleotides) difference (Supplementary Material File S3, Figs S11–S13). The Suffusa group formed a strongly supported clade with the Schweinitzii and Laurocerasi groups (BSr/PP/BSi: 90/1.0/87).
The highly supported Fraxinea group (BSr/PP/BSi: 100/1.0/99) comprises sequences representing B. fraxinea Lönnr., B. rubella, B. elongata Gerasimova & A. Beck and a new lineage. Detailed morphological analysis showed that this lineage differs from recently known species in Bacidia s. str., forming the highly supported sister group to B. elongata (BSr/PP/BSi: 99/1.0/98). Consequently, Bacidia obtecta sp. nov. is described here.
The recently described B. albogranulosa Malíček et al. nested together with B. polychroa (Th. Fr.) Körb., B. diffracta S. Ekman and B. sachalinensis Gerasimova et al. (Polychroa group) with the highest support (BSr/PP/BSi: 100/1.0/99). The sister relationship with the B. polychroa lineage (Knutsson 91-215) received high support in the RAxML phylogeny (BSr/PP/BSi: 71/-/59) but recovered a polytomy in BI.
Bacidia ekmaniana is the sister group of B. absistens and B. squamulosula, but without significant support (BSr/PP/BSi: 56/0.77/63). Together with B. arceutina, B. scopulicola (Nyl.) A. L. Sm. and B. sipmanii M. Brand et al. (Arceutina group) they formed a well-supported clade in RAxML (BSr/PP/BSi: 71/0.77/68). In the multilocus phylogeny, the clade was highly supported in BI only (BSr/PP/BSi: 66/0.97/62). Bacidia arceutina from Switzerland (LG579) formed a separate lineage to individuals from the United Kingdom (FR799125–FR799127) and Sweden (Ekman 3110), with high support in the ML phylogenies (BSr/PP/BSi: 97/0.67/97). Compared to sequences from the United Kingdom, it contained a 1% (= five nucleotides) difference in nrITS and a 1% (= six nucleotides) difference in nrLSU. Likewise, the sequence from Sweden differed by 1% (= three nucleotides) in the mtSSU compared to one from Switzerland (LG579).
The basal clade with B. thiersiana and B. hostheleoides received high support but revealed long branches (BSr/PP/BSi: 85/0.96/85).
Three main clades were recovered in Bacidia s. str.: clade I comprises the Laurocerasi, Schweinitzii and Suffusa groups; clade II includes the Fraxinea, Polychroa, and Arceutina groups, and clades of B. ekmaniana, B. absistens and B. squamulosula; clade III, sister to the others, includes B. thiersiana and B. hostheleoides.
Discussion
The multilocus phylogeny resulted in groups congruent with previous results based on nrITS (Ekman Reference Ekman2001; Gerasimova et al. Reference Gerasimova, Ezhkin and Beck2018), but significantly increased backbone support of the clades in both multilocus and all-taxa phylogenetic trees (Figs 1 & 2).
The phylogenetic trees group the taxa into two large clades: the first clade (I) includes the Laurocerasi, Schweinitzii and Suffusa groups, while the second clade (II) includes the Fraxinea, Polychroa, and Arceutina groups and clades of B. ekmaniana, B. absistens and B. squamulosula. In the phylogeny, they are represented by specimens from the temperate region and can be separated based on apothecial pigment (Table 3). The well-supported third clade (III) includes B. thiersiana and B. hostheleoides, which are widespread in south-eastern North America and the Neotropics, respectively (Malme Reference Malme1935; Ekman Reference Ekman1996; Lendemer Reference Lendemer2020).
Specimens in clade I have either dark brown, red-brown and green pigments (Laurocerasi-brown and Bagliettoa-green) or a combination of these in the upper part of the hymenium and lateral exciple (Table 3). In contrast, those in clade II have a mixture of yellow, orange and/or brown apothecial pigments (Arceutina-yellow, Polychroa-brown and Rubella-orange). An exception is a clade containing B. absistens, the pigmentation of which is highly variable (see groups below). Specimens in clade III are characterized by almost colourless or faintly and diffusely pigmented internal apothecial structures (Ekman Reference Ekman1996).
The taxon sampling in the phylogeny of Bacidia s. str. still needs to be completed; therefore, division of the clades based on the apothecial pigment is provisional. Given the available data, the main focus in this discussion is on representatives of Bacidia s. str. from the RFE. For convenience, the order of the groups corresponds to the position in the phylogenetic tree from top to bottom.
Laurocerasi group (including B. biatorina, B. kurilensis and B. laurocerasi clades)
Bacidia laurocerasi is one of the most widespread species of the group, occurring in Europe, Macaronesia, Africa, South and North America, Asia, Australia and New Zealand (Ekman Reference Ekman1996; Llop Reference Llop2007; Coppins & Aptroot Reference Coppins, Aptroot, Smith, Aptroot, Coppins, Fletcher, Gilbert, James and Wolseley2009). Bacidia biatorina is reported less frequently, and known collection localities include Europe, North America and Asia. In contrast, B. kurilensis is known only from the Kuril Islands and is considered endemic (Gerasimova et al. Reference Gerasimova, Ezhkin and Beck2018). The phylogeny contains B. biatorina from Sweden, B. laurocerasi from North America and the RFE, and B. kurilensis from the RFE.
All three taxa share Laurocerasi-brown in the exciple and upper part of hymenium (Table 3). The mixture of Laurocerasi-brown and Bagliettoa-green found in the upper part of the hymenium in B. kurilensis remains unique in this group (Gerasimova et al. Reference Gerasimova, Ezhkin and Beck2018).
All three lineages formed a strongly supported group that confirms our earlier results based on nrITS locus sequences. Two B. laurocerasi individuals formed a rather distant lineage from B. biatorina and B. kurilensis. However, as they all share the same pigment in the apothecia, we consider them to be one group.
Schweinitzii group (including B. schweinitzii and B. sorediata clades)
Bacidia schweinitzii occurs in the temperate forests of Canada around the Great Lakes and the Maritimes, in eastern Asia and the eastern parts of the USA, and the temperate region of the southern part of the RFE, in Primorskiy and Khabarovskiy Krai, and Kunashir Island (Ekman Reference Ekman1996; Lendemer et al. Reference Lendemer, Harris and Ladd2016; Gerasimova et al. Reference Gerasimova, Ezhkin and Beck2018). Bacidia sorediata is so far known only from coastal south-eastern North America, particularly the Mid-Atlantic Coastal Plain (Lendemer et al. Reference Lendemer, Harris and Ladd2016).
The high variability in morphology of B. schweinitzii s. lat. has already been discussed in previous studies (Ekman Reference Ekman1996; Lendemer et al. Reference Lendemer, Harris and Ladd2016). Two morphotypes, sorediate and esorediate, were recognized based on morphology and were subsequently shown to be concordant with the molecular results, leading to the description of three new species (Lendemer et al. Reference Lendemer, Harris and Ladd2016). It has been shown that sequences within the B. schweinitzii group formed multiple clades within B. schweinitzii s. str. (esorediate populations) and Bacidia sorediata s. lat. (sorediate morphotype). Sequences derived from sorediate populations were grouped within two strongly supported clades, sister to each other but without support. Sequences derived from esorediate specimens were recovered in a series of clades whose relationships were poorly supported. Thus, data from the nrITS and mtSSU regions appear to be insufficient to resolve the relationships between sorediate and esorediate populations with confidence.
The multilocus phylogeny presented here confirmed that B. schweinitzii is paraphyletic and contains lineages of several taxa. Nevertheless, not all branches within B. schweinitzii received high support values in both the multilocus and all-taxa phylogenies, maintaining unclear relationships within this group.
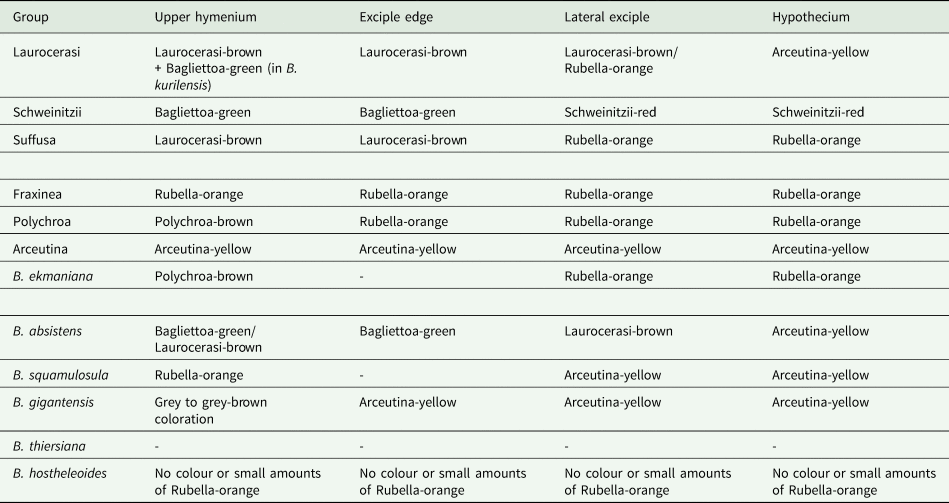
Two B. schweinitzii representatives, JG014 and JG015, form a grade and are not sister taxa in the ITS phylogeny (see Supplementary Material File S3, Figs S5–S7, available online). Instead JG015, having an abundantly granular thallus forming coral-like structures, was nested within the North American individuals. In addition, its apothecia are black, adpressed to the surface, with a brown to dark brown hypothecium merging into a darker brown exciple below and medullary part which does not form a distinct colourless zone. The spores are acicular and shorter with fewer septa, 37–46–50 × 2.5–3.5 μm, 5–7 septa (Fig. 3B & D; Supplementary Material File S4, available online). According to Lendemer et al. (Reference Lendemer, Harris and Ladd2016), such specimens can be assigned to the ‘typical’ B. schweinitzii morph by the granular thallus and apothecial coloration.

Fig. 3. Cross-sections of apothecia and thallus structure of two individuals of Bacidia schweinitzii. A & C, M-0182579 (JG014). B & D, M-0182580 (JG015). A, dark brown hypothecium, merging into the coloration of exciple below. Exciple forms a distinct colourless zone in the lateral and medullary part. B, brown hypothecium merging into the coloration of the exciple. Dark brown exciple not forming a distinct colourless zone. C, thallus smooth to warted, consisting of single or contiguous ±roundish warts. D, thallus granular, consisting of ±globose to subsquamulose granules. Scales: A & B = 200 μm; C & D = 1 mm. In colour online.
In contrast, JG014 has morphological characters that do not clearly match any species currently recognized in the B. schweinitzii group. Morphological observation demonstrated that it differs in the ±smooth to warted thallus; black caliciform apothecia, which rise above the surface; dark brown hypothecium merging into the dark brown-red exciple (K+ purplish), which forms a distinct colourless zone in the lateral part and the base of the medulla; acicular spores, 56–69–80 × 2.5–3.0 μm, 7–15 septa (Fig. 3A & C; Supplementary Material File S4). Bacidia ekmaniana also forms a distinct colourless zone in the lateral and medullary parts, in a similar way to JG014, but differs by having orange-brown apothecia and a lighter orange-brown hypothecium and exciple (K+ darker brown). It is thus likely that JG014 belongs to a different species. However, further details and careful investigation of all the type material and specimens from GenBank are necessary for making a comprehensive description and species delimitation; therefore, we currently refrain from describing new entities.
Suffusa group (including B. areolata and B. suffusa clades)
Bacidia suffusa s. lat. occurs in the eastern temperate region of North America, in the North Caucasus and the RFE (Ekman Reference Ekman1996; Gerasimova et al. Reference Gerasimova, Ezhkin and Beck2018). Bacidia areolata is known from the RFE and considered endemic. The phylogeny contains the sequences from North America and the RFE.
The specimens of B. areolata and B. suffusa formed a strongly supported clade and share Laurocerasi-brown in the exciple edge and upper part of the hymenium. Since our first molecular study, additional sequences of B. suffusa from the USA have been submitted to GenBank (AFTOL5785), including nrLSU, mtSSU and RPB1 sequences (Table 1). As the two individuals from the USA, JG080 and JG081, are represented by short sequences of nrITS2 only, the relationship to the other B. suffusa from North America remains unclear. Nevertheless, B. suffusa from the RFE formed a separate lineage from the ones collected in North America in the combined and single-locus trees based on nrLSU, mtSSU and RPB1 sequences. We could not study the individual from GenBank and suggest including more specimens from North America and other regions. Thus, we consider individuals of B. suffusa from the RFE and North America as different populations but belonging to the same species.
The newly sequenced specimen of B. areolata (JG114) represents the second occurrence of this species. Compared to the type specimen (JG037, M-0182592), it also has three layers of enlarged lumina cells along the edge of the exciple. The thallus structure, apothecial pigment and measurements are also in the range found in the type specimen. Nevertheless, JG114 differs from the type specimen by the lack of any pruina on the apothecial margin and the presence of abundant colourless crystals in the lateral exciple, dissolving in KOH with the following colour changes: green → yellow → colourless.
Fraxinea group (including B. obtecta, B. elongata, B. rubella and B. fraxinea clades)
The most widespread taxon of the group, B. rubella, has a Holarctic distribution and is known from Europe, Macaronesia, Africa, Asia and North America (Ekman Reference Ekman1996; Llop Reference Llop2007; Coppins & Aptroot Reference Coppins, Aptroot, Smith, Aptroot, Coppins, Fletcher, Gilbert, James and Wolseley2009). Bacidia fraxinea is less frequent and occurs in eastern parts of north and central Europe and the northern Mediterranean region (Ekman & Nordin Reference Ekman and Nordin1993). In contrast, both B. elongata and B. obtecta are known only from Sakhalin and can be considered endemic (Gerasimova et al. Reference Gerasimova, Ezhkin and Beck2018).
The phylogeny contains specimens of B. rubella from Europe and the RFE, B. fraxinea from Europe, and B. elongata and B. obtecta from the RFE. The phylogeny recovered the whole clade with high support, but the relationship between B. rubella and B. fraxinea remains unclear. The only sequence of B. fraxinea (Johansson 1620) was nested together with other B. rubella included in the analysis. Therefore, more specimens of B. fraxinea need to be studied.
All four taxa share Rubella-orange in the upper part of the hymenium, hypothecium and exciple. Moreover, they all have a similar coloration of the apothecia (orange to orange-brown), often with white pruina along the margin. Three taxa, B. obtecta, B. elongata and B. fraxinea, are characterized by smooth, warted to wrinkled thalli. This contrasts with the granular thallus of B. rubella.
Recent studies showed that B. elongata forms a separate entity sister to B. fraxinea, differing in its exciple structure that usually has up to four layers of enlarged lumina cells (Gerasimova et al. Reference Gerasimova, Ezhkin and Beck2018). This feature is also characteristic of the closely related and newly described B. obtecta, whereas B. fraxinea and B. rubella have only a single layer of enlarged lumina cells. Compared with B. elongata, B. obtecta has abundant colourless crystals in the upper part of the hymenium and lateral exciple, and spores with fewer septa (a detailed description is provided under Taxonomy and in Table 4).
Table 4. Main characters separating Bacidia obtecta from the closely related B. elongata and B. fraxinea. Measurements for B. fraxinea are based on those from Ekman & Nordin (Reference Ekman and Nordin1993) and measurements of B. elongata from Gerasimova et al. (Reference Gerasimova, Ezhkin and Beck2018). Quantitative information of our measurements is given as (min–) average ± SD (–max), while that for B. fraxinea was taken from the original manuscript.
Polychroa group (including B. albogranulosa, B. polychroa, B. diffracta and B. sachalinensis clades)
Bacidia polychroa represents the most widespread species of the group, occurring in Europe, North and South America, and Asia (Ekman Reference Ekman1996; Llop Reference Llop2007; Coppins & Aptroot Reference Coppins, Aptroot, Smith, Aptroot, Coppins, Fletcher, Gilbert, James and Wolseley2009). The known collection localities of B. albogranulosa include the Czech Republic, Poland, Russia (Caucasus) and Ukraine (Malíček et al. Reference Malíček, Palice, Vondrák, Łubek and Kukwa2018). Bacidia diffracta and B. sachalinensis are endemics and are so far known from North America and the RFE, respectively (Ekman Reference Ekman1996; Gerasimova et al. Reference Gerasimova, Ezhkin and Beck2018). The phylogeny contains B. polychroa from Sweden, B. albogranulosa from Europe, B. diffracta from North America, and B. sachalinensis from the RFE.
The phylogeny recovered the whole clade with high support and is concordant with the previous results of Gerasimova et al. (Reference Gerasimova, Ezhkin and Beck2018), who provide a detailed discussion. All taxa within the clade share Polychroa-brown in the hypothecium, exciple and hymenium, the K+ purplish reaction in apothecial cross-sections. However, this reaction is unknown for the recently described B. albogranulosa since it is known only in its sterile form (Malíček et al. Reference Malíček, Palice, Vondrák, Łubek and Kukwa2018).
Arceutina group (including B. arceutina, B. scopulicola and B. sipmanii clades)
Bacidia arceutina is the most widespread epiphytic species of the group. The known collection localities include Europe, Asia and North America (Coppins & Aptroot Reference Coppins, Aptroot, Smith, Aptroot, Coppins, Fletcher, Gilbert, James and Wolseley2009). In contrast, B. scopulicola and B. sipmanii are almost exclusively epilithic, occurring in coastal Europe from Scandinavia to the Azores, and in North America (Coppins & Aptroot Reference Coppins, Aptroot, Smith, Aptroot, Coppins, Fletcher, Gilbert, James and Wolseley2009). The phylogeny includes the specimens of all three taxa collected in Europe.
All three taxa share Arceutina-yellow in the apothecial structures. So far, none of the three species have been observed from the RFE. Bacidia arceutina from Switzerland formed separate lineages from those originating in the United Kingdom (FR799125-FR799127) and Sweden (Ekman 3110). However, given the low genetic differentiation in nrITS, nrLSU and mtSSU (1% in each), we regard them as conspecific.
Clades including B. ekmaniana, B. absistens and B. squamulosula
Bacidia ekmaniana is known from south-eastern North America, occurring from the Appalachian Mountains to the Coastal Plain and southern Interior Highlands (Lendemer et al. Reference Lendemer, Harris and Ladd2016). The known distribution of B. absistens includes Europe, Macaronesia, Africa, Asia and North America (Ekman Reference Ekman1996; Coppins & Aptroot Reference Coppins, Aptroot, Smith, Aptroot, Coppins, Fletcher, Gilbert, James and Wolseley2009; Gerasimova Reference Gerasimova2016), whereas B. squamulosula is known from South and North America (Dahl Reference Dahl2017; McMullin et al. Reference McMullin, McCune and Lendemer2020).
Bacidia ekmaniana and B. squamulosula share a mixture of Rubella-orange and Arceutina-yellow in the apothecia, a feature they share with other taxa from clade II (Table 3). In contrast, the pigmentation in B. absistens is highly variable. It may contain a mixture of Bagliettoa-green, Laurocerasi-brown or grey coloration in the upper hymenium and exciple edge (Gerasimova Reference Gerasimova2016). Thus, the pigmentation of B. absistens is unique in this clade. The significant morphological variation in this species requires further study, involving more specimens.
The recently published B. gigantensis formed a sister clade to B. absistens and B. squamulosula in the original publication of McMullin et al. (Reference McMullin, McCune and Lendemer2020) and the relationships of these taxa are discussed in detail there. Bacidia gigantensis has grey to grey-brown coloration in the upper hymenium and exciple, also observed in B. absistens (Gerasimova Reference Gerasimova2016). The three taxa, B. absistens, B. gigantensis and B. squamulosula contain secondary compounds, 4-O-methylcryptochlorophaeic and homosekikaic acids. This is unusual and is not known for any other Bacidia s. str., which contain atranorin as the main secondary compound (Ekman Reference Ekman1996). In addition, all three taxa are characterized as having minute colourless crystals in the exciple.
Clade including B. thiersiana and B. hostheleoides
Bacidia thiersiana (deposited as Bacidia lutescens in GenBank) is widespread in south-eastern North America, while B. hostheleoides is widely distributed in the Neotropics (Malme Reference Malme1935; Ekman Reference Ekman1996; Lendemer Reference Lendemer2020). Bacidia thiersiana is characterized by having some oil droplets in the hypothecium and by the production of lobaric acid, both unique characters in Bacidia s. str. phylogeny. Both taxa are characterized by almost colourless or faintly and diffusely pigmented internal apothecial structures (Ekman Reference Ekman1996; Lendemer Reference Lendemer2020). However, B. hostheleoides has small amounts of Rubella-orange in the proper exciple, hypothecium and hymenium when pigmented (Ekman Reference Ekman1996).
These taxa belong to a well-supported clade with long branches, most likely resulting from incomplete species sampling rather than a genuine relationship. Therefore, further sampling in this group is still required to draw reliable conclusions.
Taxonomy
Bacidia obtecta Gerasimova, A. Ezhkin & A. Beck sp. nov.
MycoBank No.: MB 838949
Similar to Bacidia elongata but differs by abundant colourless crystals in the upper part of the hymenium and lateral exciple, dissolving in KOH, and by having spores with fewer septa.
Type: Russia, Sakhalin, Tymovskiy District, valley of the Tym River, Zonal'noye village surroundings, 51°04′42.6899″N, 142°44′25.8756″E, c. 100 m a.s.l., floodplain forest, on the bark of Populus maximowiczii, 4 June 2017, A. K. Ezhkin [B4/02.2018] (M M-0308496—holotype; UPS, SAK 1368—isotypes).
(Fig. 4)
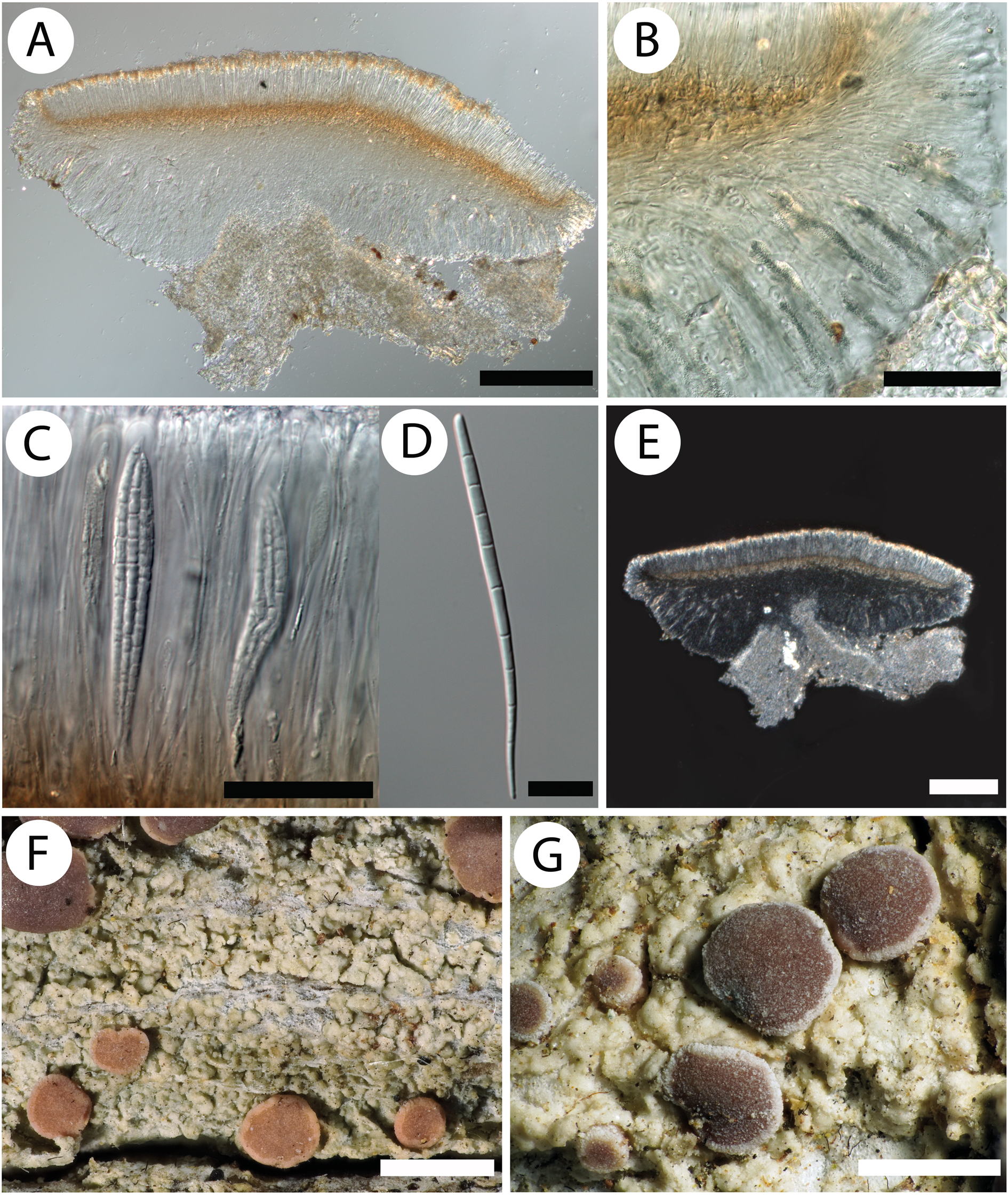
Fig. 4. Cross-section of apothecium and thallus structure of Bacidia obtecta (M-0308496, holotype). A, cross-section of apothecium. B, clusters of crystals radially arranged in the lateral part of the exciple. C, asci with ascospores. D, acicular multiseptate ascospore. E, crystals in the cross-section of apothecium visualized using polarized light. F, general overview of apothecia and thallus structure. G, detailed view of apothecia and thallus structure. Thallus wrinkled, irregularly shaped. Rusty brown apothecia with scurfy surface and white pruina along the margin. Scales: A, B & E = 200 μm; C = 50 μm; D = 20 μm; F & G = 1 mm. In colour online.
Thallus indefinite, continuous or discrete, relatively thick, wrinkled, sinuous, consisting of scattered or continuous wrinkled, irregularly shaped or ±convex warts, grey, grey-green to yellowish green. Prothallus white. Photobiont chlorococcoid.
Apothecia (0.4–)0.85 ± 0.25(–1.5) mm, ±plane when young, soon becoming markedly convex, then margin is excluded; disc pruinose, with scurfy/incrusted surface; margin of young and medium-aged apothecia often with thick white pruina. Disc beige, pale orange, orange-brown to rusty brown. Margin distinct, always paler than disc, beige or pale brown. Epithecium colourless, interspersed with abundant colourless crystals between the paraphysis apices, less than 1 μm. Exciple laterally 86–94.3–98 μm high, with abundant colourless crystals, less than 1 μm. Rim pale to intense straw-coloured, along the edge with up to four layers of enlarged lumina cells that are 3–5 μm wide and 5–13 μm long. Lateral part pale to intense straw-coloured or yellow. Medullary part of the same colour as lateral part, downwards almost colourless. Hymenium 73.5–108–135 μm high. Hypothecium pale to intense orange-brown (Rubella-orange). Paraphyses simple, thin, colourless, sometimes bifurcate, 1.0–1.5 μm wide, ±clavate or only slightly swollen at the apices, 1.5–3.0 μm, without internal pigment. Ascospores acicular, straight, (47–)61.2 ± 7.0(–79) μm long (n 1 = 88, n 2 = 3), (2.0–)3.0 ± 0.36(–4.0) μm wide (n 1 = 88, n 2 = 3), with (1–)6 ± 2(–11) septa (n 1 = 88, n 2 = 3).
Pycnidia not observed.
Pigments
Hypothecium K+ intensifying or yellowish, exciple C+ and N+ yellow, subsequently becoming colourless. Rubella-orange in hypothecium and exciple. Crystals dissolve in K, forming the following reaction: green → yellow → colourless; do not dissolve in N or C.
Etymology
Apothecial disc covered with pruina on top, which gives a ‘powdery’ appearance.
Habitat and distribution
The specimens were collected in the floodplain forest in river valleys on the bark of Chosenia arbutifolia and Populus maximowiczii. So far, known only from Sakhalin. Probably endemic to the island.
Comments
Bacidia obtecta is similar to B. elongata and B. fraxinea but differs in several distinct features, summarized in Table 4. Compared to B. elongata and B. fraxinea, it is distinguished by the apothecial colour, which is mainly brown to rusty brown, abundant colourless crystals in the upper part of the hymenium and lateral exciple, which dissolve in KOH but not in HNO3, and spores with fewer septa. From B. fraxinea it can be additionally distinguished by the number of enlarged lumina cells along the exciple edge.
Additional specimens examined
Russia: Sakhalin: Tymovskiy District, valley of Pilenga River, Adotymovo village surroundings, 51°02′07.6236″N, 142°49′26.5692″E, c. 160 m a.s.l, floodplain forest, on bark of Populus maximowiczii, 5 vi 2017, A. K. Ezhkin [B2/02.2018; GPS 547] (M M-0308497, SAK 1365); ibid., valley of Tym River, Zonal'noye village surroundings, 50°38′51.2951″N, 142°45′48.2868″E, c. 160 m a.s.l., floodplain forest, on bark of Chosenia arbutifolia, 6 vi 2017, A. K. Ezhkin [B1/02.2018; GPS 551] (M-0308498, SAK 1362).
Acknowledgements and Author Contribution
We thank the associate editor and two anonymous reviewers for their highly valuable comments and improvements. We are grateful to David Richardson (Halifax, Canada) for his corrections to the English text. JG was supported by a BAYHOST fellowship from the Bayerische Staatsministerium für Bildung und Kultus, Wissenschaft und Kunst. Molecular work was supported by a grant to AB from the Bayerische Staatsministerium für Bildung und Kultus, Wissenschaft und Kunst within the ‘Barcoding Fauna Bavarica’ framework. The Russian Foundation for Basic Research partially supported the work of AE (No. 18-04-00098 A). AB and JG designed the study and wrote the paper; JG performed the experiments; AE and ED provided specimens and approved the manuscript.
Author ORCIDs
Julia V. Gerasimova, 0000-0002-3212-3596; Aleksandr Ezhkin, 0000-0002-2242-2250; Andreas Beck, 0000-0003-2875-4464.
Supplementary Material
To view Supplementary Material for this article, please visit https://doi.org/10.1017/S0024282921000396











