In 2009, the Osaka University Center for Twin Research (OUCTR) was established as an affiliated research institute of Osaka University Graduate School of Medicine (Dean: Professor Eiichi Morii), and the main office is located in the Division of Health Sciences, Osaka University Graduate School of Medicine (Division Director: Professor Yuko Ohno). Research activities at OUCTR are based on the current Osaka University Twin Registry, which was founded in the 1980s and is considered to be one of the largest twin registries in Japan (Hayakawa et al., Reference Hayakawa, Shimizu, Kato, Onoi and Kobayashi2002; Hayakawa, Kato, Onoi, Hayashi et al., Reference Hayakawa, Kato, Onoi, Hayashi, Cai, Kanamori and Kadota2006; Hayakawa, Kato, Onoi, Cai et al., Reference Hayakawa, Kato, Onoi, Cai, Kanamori, Doi and Kadota2006; Hayakawa et al., Reference Hayakawa and Iwatani2013).
With Professor Norio Sakai as director and Professors Masahiko Koizumi and Masanori Takahashi as vice directors, the OUCTR consists of the following four departments: Department of Ethics and Data Management (Chief: Professor Hiroyasu Iso), Department of Bioresources (Chief: Professor Masanori Takahashi), Department of Research Infrastructure (Chief: Professor Norio Sakai) and Department of Research Promotion (Chief: Professor Mikio Watanabe).
One of the strengths of the OUCTR is that it is a multidisciplinary organization comprising experts in a wide variety of specialties from the natural and social sciences of Osaka University. Its uniqueness has been highly recognized by the Japanese Ministry of Education, Culture, Sports, Science and Technology (MEXT), and the university’s administration and OUCTR has been financially supported by Osaka University and MEXT’s National University Operation Subsidy since 2011 for three consecutive funding terms. The specific aims vary with each term, but OUCTR’s principal goal has been consolidation of the twin research infrastructure, by collecting and storing diverse twin research data for current and future research projects. Therefore, we have been conducting a wide range of data collection on twins and have established a biobank and databases, based in the Osaka University Twin Registry.
Osaka University Twin Registry
The history of the Osaka University Twin Registry (hereafter referred to as the Registry) dates back to the 1980s when the first director (Dr. Hayakawa) started a registry that consisted mainly of older twins. The Registry has subsequently been expanded to include twins of all ages from all over Japan, with about 3000 active registrants. However, the Registry still includes a substantial number of older twins. Twins under 18 years comprise about 20% of the registrants, and almost 60% of the adult twin registrants are aged over 60 years.
The Registry is volunteer-based and open to new enrollment at all times from any part of Japan, although most new registrants are from Osaka and the surrounding regions (Central Japan). Accessing the birth registry is very difficult in Japan, and thus we have relied on voluntary enrollment from twins. Our efforts to increase new enrollment include advertising in newspapers, distributing posters and leaflets to neighborhoods and organizing public lectures and symposiums. As a result, we have maintained the registry size even with considerable attrition owing to death among the older registrants.
Participants in the Core Survey
There are several separate surveys and projects in progress at OUCTR, but the core survey is a pillar of all projects and is described here.
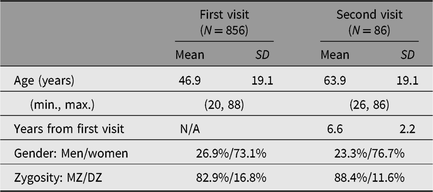
The core survey aims to conduct comprehensive data collection from twins of the Registry and to store bioresources and information in databases. We send letters by post to pairs of registered twins (aged 20 years or older) from all over Japan to recruit survey participants. When both twins of a pair agree to participate, we invite them to Osaka University, which is located in Central Japan, or to a location set up near their residence. Participants then undergo a one-day or half-day comprehensive survey comprising a physical examination, physiological tests and epidemiological questionnaires. As of June 1, 2019, a cumulative total of over 450 twin pairs have participated in the core survey. This includes twins participating in the longitudinal survey who attended their second visit after five years. The mean ages of the participants are 46.9 years (age range: 20–88) and 63.9 years (age range: 26–86) for the first visit and second visit, respectively. Of the pairs, 82.9% are monozygotic (MZ) and 16.8% are dizygotic (DZ) twins (one pair refused to get tested for zygosity). Of the participants, 73.1% are women and 26.9% are men. Descriptive statistics of the participants are shown in Table 1.
Table 1. Descriptive statistics of the core survey
Data Collection
We collect physical data (e.g., height, body weight, blood pressure, theoretical visceral fat, pulse wave velocity and bone density); epidemiological data (e.g., medical history, lifestyle, quality of life, mood status, cognitive function and nutritional status); electrocardiography, ultrasonography (carotid artery and thyroid); dentistry, dermatological assessment; positron emission tomography (PET); magneto-encephalogram (MEG); brain magnetic resonance imaging (MRI); and functional MRI (fMRI). These data are then aggregated and systematically stored in specific databases.
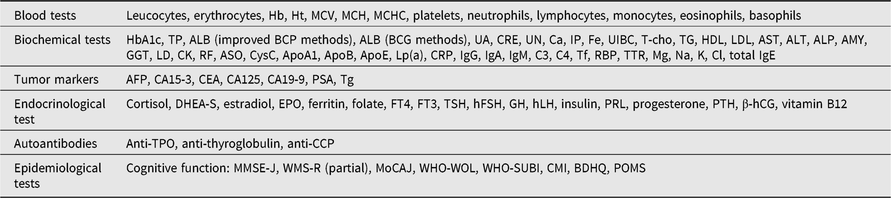
We also obtain peripheral blood from the participants. Genomic DNA is extracted, serum is separated from the blood, whole blood cells are lysed for isolation of total RNA, and the samples are preserved in deep freezers for future analyses. We perform the laboratory tests listed in Table 2 to assess the clinical condition of each participant.
Table 2. Laboratory tests and epidemiological tests performed in the OUCTR
All data collection has been approved by the Research Ethics Committee of Osaka University, and written informed consent was obtained from all participants in the core survey.
Zygosity Determination
Zygosity determination of the twins is based on the 15-short tandem repeat (STR) loci using PowePlex 16® System (Promega, Madison, WI). A twin pair with perfect matching of the STRs is classified as MZ.
Research Projects Using the Core Survey Data
Genetic/Epigenetic Study
At OUCTR, genetic/epigenetic information and bioresources, including RNA, have been collected to analyze the detailed mechanisms of how various environmental factors affect gene expression. Whereas genomic DNA remain unchanged throughout a person’s lifetime, epigenetic modifications are possible and are thought to be associated with the fact that some MZ twins have different phenotypes. In addition, it has recently been understood that micro-RNA and other factors that affect gene expression are crucial for the epigenetic regulation of gene expression. As a result, understanding epigenetic regulation and the ways in which various environmental factors affect epigenetic modification are extremely important, making this an integral area of the twin research infrastructure of the OUCTR.
The ultimate goal of the OUCTR is to develop ways to identify environmental factors that affect the disease phenotypes of an individual by taking into account the individual’s genetic background. Analyzing how environmental factors affect diseases and the expression of genes from an epigenetic perspective will enable the utilization of genetics to estimate the prognosis, such as the onset and progression of diseases. This will in turn provide individuals with adequate preventive measures and personalized instructions for modifying their daily habits.
To achieve these goals, the OUCTR has set up a biobank to store DNA, RNA, and serum as well as databases with analyzed genetic/epigenetic data and associated information. Genomic single nucleotide polymorphism (SNP) genotyping is performed using the Illumina® Infinium HumanOmni5-Quad BeadChips (Illumina, San Diego, CA). Methylation levels are quantified using the Infinium HumanMethylation450 BeadChip Kit, which was recently updated to the MethylationEPIC BeadChip (Illumina) according to the manufacturer’s standard protocol, yielding over 450,000 and 850,000 methylation sites at single-nucleotide resolution for each sample, respectively. Furthermore, we are planning to conduct transciptome analyses using the stored RNA and have conducted pilot analyses on several samples.
At present, several projects are ongoing in this area, and some of the findings have been published elsewhere and more are on the way (Watanabe, Honda et al., Reference Watanabe, Honda and Iwatani2016; Watanabe et al., Reference Watanabe, Takenaka, Honda and Iwatani2018).
Epidemiology and Physiological Assessment
We continue to collect epidemiological information to clarify lifestyle and environmental factors giving rise to phenotypic differences among twins. As part of the epidemiological data collection, we are using validated Japanese versions of a series of questionnaires; for example, Cornell Medical Index (CMI; Kanehisa & Fukamachi, Reference Kanehisa and Fukamachi1983) for general medical conditions, Brief-type Self-administered Diet History Questionnaire (BDHQ; Kobayashi et al., Reference Kobayashi, Murakami, Sasaki, Okubo, Hirota, Notsu and Date2011; Sasaki et al., Reference Sasaki, Yanagibori and Amano1998) for dietary conditions, Mini-Mental State Examination (MMSE-J; Sugishita et al., Reference Sugishita, Hemmi and Iwatsubo2010), Wechsler Memory Scale Revised (WMS-R; Matsuda et al., Reference Matsuda, Saito and Sugishita1998) and Japanese version of Montreal Cognitive Assessment (MOoCA-J; Fujiwara et al., Reference Fujiwara, Suzuki, Yasunaga, Sugiyama, Ijuin, Sakuma and Ishii2010) for cognitive ability, Profiles of Mood States (POMS; Yokoyama et al., Reference Yokoyama, Araki and Kawakami1990) for emotional states, WHO-QOL 26 (Tazaki & Nakane, Reference Tazaki and Nakane1997) for quality of life and WHO-SUBI (Ohno & Yoshimura, Reference Ohno and Yoshimura2001) for subjective well-being. Anthropometric measurements, such as height, weight and waist circumference, and basic physical functions, such as blood pressure, grip strength and hearing level, are also collected. Various physiological tests provide further information, including theoretical visceral fat, bone density, cardiac function by electrocardiography (ECG), body composition and ultrasonographic images (carotid artery and thyroid). Data availability of each test may vary among participants.
Dentistry
Data collection is conducted by collaborating dentists affiliated with the OUCTR as a part of the comprehensive physical examination. Dental and periodontal examinations were conducted to evaluate the dental condition of the participants. In addition, salivary flow, maximum occlusal force and masticatory performance are measured. In the past, orthopantomography and dental impressions were also collected. Furthermore, we use the Japanese versions of Oral Health Impact Profile (OHIP-14; Ikebe et al., Reference Ikebe, Watkins, Ettinger, Sajima and Nokubi2004) and OHIP-EDENT (Sato et al., Reference Sato, Kaiba, Yamaga and Minakuchi2012) to measure oral health-associated quality of life. Findings from the dental study have been reported elsewhere (Kurushima et al., Reference Kurushima, Ikebe, Matsuda, Enoki, Ogata, Yamashita and Maeda2015a, Reference Kurushima, Ikebe, Matsuda, Enoki, Ogata, Yamashita and Maeda2015b).
Microbiome Study
Microbiome analyses have been performed as a new approach to understanding disease-environment relationships. The human gut microbiome has been studied extensively to show the link between health and diseases, and is considered to be closely involved in human function, including metabolism, nutrition and immune system. Certain conditions of gut microbiota have been linked with various diseases. In addition, as the microbiome is affected by race/ethnicity and lifestyle habits, research on the Japanese population is necessary to uncover the unique effect of Japanese lifestyle on the microbiome. The OUCTR has collected fecal samples from more than 100 twins and will collect more from various participants to better understand the human gut microbiome.
The collected fecal samples are mixed well and stored in vials with Guanidine thiocyanate (GuSCN) solution (TechnoSuruga Laboratory Co., Ltd., Shizuoka, Japan) at 4 C until DNA extraction. DNA is extracted using a Gene Prep Star PI-80X device (Kurabo Industries Ltd.) and the V3–V4 region of the 16S rRNA gene is amplified and sequenced using Illumina MiSeq. The whole process of fecal sample analysis is conducted according to the protocol described by Hosomi et al. (Reference Hosomi, Ohno, Murakami, Natsume-Kitatani, Tanisawa, Hirata and Kunisawa2017). In addition, saliva was collected from a limited number of participants.
Functional MRI Study
Whereas genetic contribution has been reported in various types of cognitive functions, little is known about how genetic and environmental factors affect individual differences of perception and recognition, and thus its effects on the representation of natural experiences are yet to be investigated. Whole-brain activity is measured using fMRI in twin participants who are made to watch 70 min of natural audio-visual movies consisting of a variety of complex scenes to induce perception and cognition activities in daily life situations. Study results are in process for publication.
Past Projects
Projects described here are not being conducted at the time of submission, but data are available for analyses jointly with the rest.
Neurology
We measured brain functions using MEG and brain MRI. MEG was performed with visual stimuli using a 160-channel whole-head neuromagnetometer device (MEG vision NEO; Ricoh Company, Ltd., Yokohama, Japan). MEG data were recorded using an online band-pass filter between 0.01 and 200 Hz and sampled at 1000 Hz. Findings from the MEG study and details have been reported elsewhere (Araki et al., Reference Araki, Hirata, Sugata, Yanagisawa, Onishi, Watanabe and Yorifuji2014, Reference Araki, Hirata, Yanagisawa, Sugata, Onishi, Watanabe and Iwatani2016). MRI images were obtained using two different 3.0-Tesla high-resolution scanners, mainly to align the MEG data. In addition, volume measurements were analyzed and reported high heritability of brain volumes (Lukies et al., Reference Lukies, Watanabe, Tanaka, Takahashi, Ogata and Omura2017).
Nuclear medicine
The PET technique visualizes physical functions in vivo by detecting positron-emitting radioisotopes. We used 18F-fluorodeoxyglucose (18F-FDG) to visualize the distribution of glucose metabolism. Our target organs are from the brain to the femoral region, and the levels of FDG accumulation in the target organs were evaluated. To our knowledge, only a few researchers have reported twin studies using FDG-PET. Similarities and differences in FDG accumulation between twin pairs were analyzed (Watanabe, Kato et al., Reference Watanabe, Kato, Shimosegawa and Hatazawa2013, Reference Watanabe, Kato, Shimosegawa and Hatazawa2016).
Dermatological study
To contribute to the research of anti-aging and improved quality of life, facial skin measurements using specific imaging techniques were conducted. Participants were asked to answer a questionnaire about lifestyle relevant to skin aging, such as sun exposure and skin care. Furthermore, the participants’ faces were assessed by an imaging device, to analyze spots and redness on their facial skin, enabling us to measure skin temperature, moisture content, fat quantity, viscoelasticity and the stratum corneum. The results of the similarities and differences found are reported elsewhere (Ichibori et al., Reference Ichibori, Fujiwara, Tanigawa, Kanazawa, Shingaki, Torii and Hosokawa2014).
Future Plans
In 2019, as we open the third term of OUCTR, we plan to expand our survey to pediatric twins on a larger scale in addition to the present adult twin projects. We will work in collaboration with large-scale, non-twin familial cohort studies in Japan, to obtain perinatal/postnatal information on pediatric twins. Furthermore, we will start a new pediatric twin survey with support from local women’s and children’s hospitals, focusing on the developmental origin of health and diseases.
As there are few large-scale twin studies in Japan, it is very important to work collaboratively. Further collaborations will reach out to twin research institutions inside and outside of Japan for comparative studies, particularly in the field of genetics/epigenetics and microbiome studies.
Acknowledgments
We thank all twins in the Registry for invaluable contribution to our research. The Osaka University Center for Twin Research is supported by grants from the Ministry of Education, Culture, Sports, Science and Technology. We are also thankful to Beckman Coulter, Inc. (Tokyo, Japan) for their collaborative work. We would also like to thank Kanako Akada, Miho Anayama, Rie Honda, Kazuya Tsubaki and Noriko Ushio for administrative assistance.
Osaka Twin Research Group
Norio Sakai, Masanori Takahashi, Masahiko Koizumi, Kei Kamide, Shinji Kihara, Hiroko Watanabe, Mikio Watanabe, Chika Honda and Rie Tomizawa.




