INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) is well established as a major hospital-associated pathogen that has spread into the community through discharge of colonized patients. Recently, however, MRSA has increasingly been described in patients in the community who lack the obvious risk factors for acquisition of hospital-acquired MRSA (HA-MRSA) [Reference Herold1, Reference Naimi2]. While most of these community-associated MRSA (CA-MRSA) infections have been associated with relatively mild, often self-limiting, skin and soft-tissue infection, they can be highly contagious and cases of severe necrotizing soft-tissue and lung infections have been reported [3, Reference Mongkolrattanothai4].
It is now believed that these CA-MRSA strains have evolved separately from typical nosocomial strains. Such strains are usually susceptible to many non-β-lactam antibiotics and molecular typing suggests they are no more closely related to HA-MRSA than to methicillin-sensitive Staphylococcus aureus (MSSA) [Reference Enright5]. Most CA-MRSA strains that have been investigated possess the small (21–24 kb), mobile staphylococcal chromosomal cassette mec type IV (SCCmec IV). In addition, many CA-MRSA have recognizable virulence factors, often the Panton–Valentine leukocidin (PVL) gene, which makes them of epidemiological and clinical significance.
Strains of CA-MRSA have recently been reported as causing sporadic infections in hospitalized neonates and more rarely outbreaks of infection [Reference Bratu6–Reference Sax8]. These strains carried the PVL gene. We describe an outbreak in a nursery and maternity unit involving an ST5, SCCmec IV strain of CA-MRSA carrying an exfoliative toxin and an enterotoxin. The prevalence of this strain, (ST5-MRSA-IV), in Scotland is also described.
MATERIALS AND METHODS
Outbreak investigation and management
From 23 September 2003 to 11 March 2004 a cluster of skin and soft-tissue infections due to MRSA involved neonatal and maternity patients in a Grampian community hospital 30 miles north of Aberdeen. The 43-bed hospital has a labour and delivery unit of one bed and a unit of seven beds, including three single rooms, that houses healthy newborns and maternity patients. Individual healthcare workers typically work on all of these units.
After recognition of the outbreak, nursing and medical staff were reminded of the ways staphylococci spread and contact precautions were reinforced. The availability of alcohol hand gel was increased and the importance of hand washing stressed. Staff uniform and finger and thumb prints were taken by plate impression onto horse blood agar. Environmental and equipment swabs included vents, fans, baths, cord clamps, thermometers, mattresses, blood pressure cuffs, stethoscopes, chairs and telephones. Mother and baby swabs were taken from anterior nares, axilla, throat, groin, any wounds and from the umbilicus of neonates. The noses and throats of staff were swabbed. Environmental and body-site sampling was by Copan charcoal swabs (Sterilin, UK) moistened in sterile water. Screening swabs were collected between 3 December 2003 and 6 April 2004.
Local doctors including paediatricians were alerted to the problem. Cases were defined as MRSA infections in patients from the labour, delivery and maternity units of any hospital in Grampian from September 2003 to September 2004. The medical records of all patients were reviewed to establish history, predisposing factors, treatment and outcome.
Laboratory methods
Swabs were plated directly onto ORSAB agar (Oxoid, UK). Colonies resembling MRSA were identified by standard techniques. Susceptibility testing was by Clinical Laboratory Standards Institute (CLSI) agar disc diffusion. In addition to tests carried out in the Diagnostic Laboratory, all MRSA isolates were sent to the Scottish MRSA Reference Laboratory where they were retested by Vitek (bioMérieux, France) for sensitivity to a standard range of antibiotics, subjected to urease, Tween hydrolysis and pigmentation tests, to PCR for a range of toxin genes and to typing by pulsed-field gel electrophoresis (PFGE) [Reference MacKenzie9]. The toxin genes sought by PCR were enterotoxins A, B, C, D, E, toxic shock toxin [Reference Johnson10, Reference Becker, Roth and Peters11], exfoliative toxins A and B [Reference Sakurai, Suzuki and Machida12], and PVL [Reference Lina13]. Multilocus sequence typing (MLST) (http://www.mlst.net), PCR for the mecA gene [Reference Brakstad, Aasbakk and Maeland14, Reference Bignardi15] and SCCmec typing, using the primer set described by Oliveira & de Lencastre [Reference Oliveira and de Lencastre16], was performed on selected isolates.
Surveillance study
The Scottish MRSA Reference Laboratory has typed MRSA isolates submitted from all parts of Scotland since 1997 and all new isolates from Grampian have been submitted since 2001. The typing methods used have changed with time but all isolates have had their susceptibility to a standard set of antibiotics tested and all isolates, other than some recognized phenotypically as EMRSA15 or EMRSA16, have been typed by PFGE. Since 2003, selected isolates have been typed by MLST and subjected to toxin and SCCmec typing as described above. Criteria for submission of isolates are not the same in all Scottish diagnostic laboratories.
RESULTS
Outbreak investigation and management
Between 2 October 2003 and 11 March 2004, seven neonates and one mother, all previously well, had MRSA infection of skin and soft tissue. Clinical details and timelines are shown in Table 1 (a, b). None of the patients were seriously ill and the clinical condition necessitated the use of systemic antibiotics in only one case. None required surgical drainage. In all cases the patient had been discharged before MRSA was identified. The adult who had infection of her caesarean section wound had been transferred to the main maternity hospital in Aberdeen but was readmitted to the community hospital for a day before being discharged home. Infection developed subsequent to discharge. Her baby was not infected and umbilical, nasal, throat, axilla and groin swabs did not detect MRSA. The mothers of five of the other affected babies were screened for MRSA but all were negative in nose, throat, axilla, groin and high vaginal swab. None had relevant past hospital exposure, recognized predisposing medical conditions for MRSA carriage or had received prior antibiotic therapy (except for two mothers during gestation). No MRSA infection had been reported from the unit in the previous 12 months and no other cases have occurred in the unit up to the time of writing (July 2006). No additional cases were reported in surrounding hospitals.
Table 1a. Details of outbreak cases (seven affected neonates and one mother, case 5, whose baby was not affected)
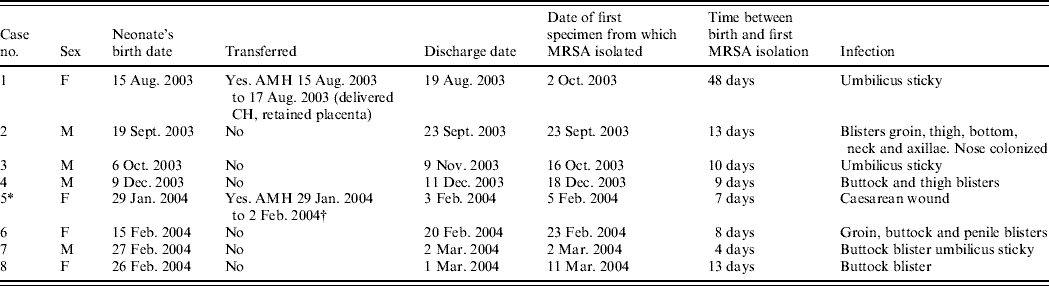
AMH, Aberdeen Maternity Hospital; FCH, Fraserburgh Community Hospital.
* Positive mother (baby negative contact screen).
† Caesarean section because of failed progression.
Table 1b. Details of outbreak cases
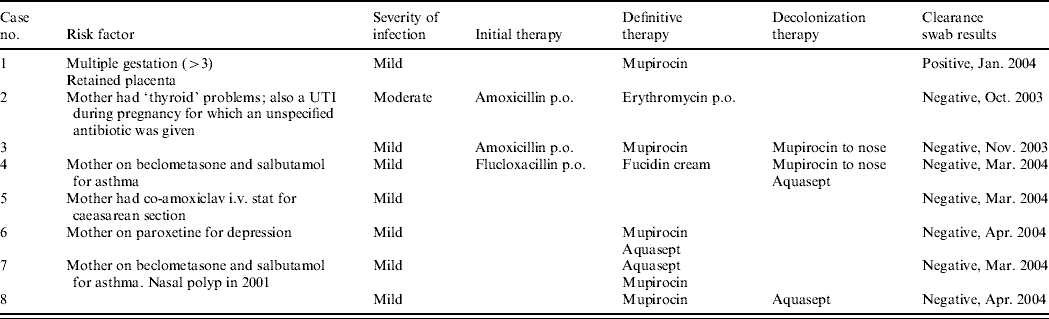
UTI, Urinary tract infection.
Susceptibility testing showed that all eight isolates were susceptible to trimethoprim, sulfamethoxazole, erythromycin, clindamycin, ciprofloxacin, rifampicin, doxycycline, chloramphenicol, linezolid, vancomycin, teicoplanin, synercid, gentamicin but resistant to fusidic acid. All isolates had one of two closely related PFGE patterns (see Fig. 1), recognized by the Reference Laboratory as associated with ST5-MRSA-IV (USA800). The SCCmec type was confirmed for three isolates and the MLST type for one isolate of each of the two PFGE patterns. All eight isolates carried the exfoliative toxin A (ETA) and enterotoxin D genes but no other toxin genes. The toxin gene tests were initially carried out because the clinical details supplied with one isolate (‘blister’) suggested that an exfoliative toxin might be involved.

Fig. 1. PFGE patterns of outbreak isolates. Key: isolates in tracks numbered from left are outbreak isolates from patients in Table 1 except where indicated. Not Ob, not part of the outbreak but a related isolate; ETA, exfoliative toxin A; SED, enterotoxin D; PVL, Panton–Valentine leukocidin. Lanes: 1, Standard (NCTC 8325); 2, ETA+, SED+, PVL−; 3, ETA+, SED+, PVL−; 4, ETA+, SED+, PVL−; 5, ETA+, SED+, PVL−; 6, ETA+, SED+, PVL− (during storage a band in the original PFGE was lost); 7, Standard (NCTC 8325); 8, ETA+, SED+, PVL−; 9, ETA+, SED+, PVL−; 10, ETA+, SED+, PVL−; 11, Comparator (ST5-MRSA-II); 12, Not Ob ETA−, SED+, PVL−; 13, Not Ob ETA+, SED+, PVL−; 14, Not Ob ETA−, SED+, PVL+.
Nineteen nursing staff worked regularly on the unit. Twelve had been on shifts when the infected patients had been in hospital and all were negative for MRSA. The uniforms of seven were cultured and were also negative as were the hands of a radiographer and a cleaner. Swabs from 45 environmental and equipment sites were cultured but all were negative for MRSA.
Surveillance study
Almost 6000 index case clinical and screening isolates of MRSA from patients in Grampian were isolated between 1997 and 2004 and referred to the Reference Laboratory. Most of the isolates were EMRSA15 (ST22-MRSA-IV, 60%) or EMRSA16 (ST36-MRSA-II, 30%), the common hospital-associated strains in Scotland as in the rest of the UK. There were 95 isolates with the same phenotype (antibiogram and biotype) and genotype (a closely related PFGE pattern) as the outbreak isolates. Neither this phenotype nor this genotype are common among referred isolates and it is probable that most of these isolates are ST5-MRSA-IV and that those that are not belong to the same MLST clonal complex. This inference is strengthened by the finding that all these isolates were positive by PCR for enteroxin D; however, only 60% of them carried the ETA gene.
Minor variations seen in PFGE pattern did not correlate completely with the presence or absence of the ETA gene. Five other infants (aged <1 year) yielded ST5-MRSA-IV strains during this period but only three were positive for ETA and all five cases were sporadic. During this period 14 PVL-positive MRSA isolates from Grampian were recognized but none of these were ST5 (seven were ST80-MRSA-IV and seven were ST8-MRSA-IV) and no outbreaks were recognized. The outbreak strain (ST5-MRSA-IV, ETA positive) appeared unique to Grampian during this period but some 20 isolates with similar phenotypes and closely related PFGE patterns have been received by the Reference Laboratory since 1997 from other Scottish laboratories. Not all such isolates received before 2003 were tested for toxins but all of the eight tested were negative for the ETA gene (although three were positive for the enterotoxin D gene) and no outbreaks have been recognized. In 2005 a PVL-positive (ETA-negative) isolate, inferred from its phenotype and PFGE pattern to be ST5-MRSA-IV, isolated from a patient in Scotland was referred to the Reference Laboratory but its PFGE pattern was slightly different from the two outbreak patterns (see Fig. 1) and it did not carry the enterotoxin D gene.
DISCUSSION
We describe the healthcare transmission of CA-MRSA strain ST5-MRSA-IV (ETA positive) in previously healthy neonates and a previously healthy mother who had undergone a caesarean section. All but one baby developed infection within 2 weeks of birth and so probably acquired the MRSA in the hospital; however, no environmental or staff source could be found so the origin and mechanisms of transfer have not been established. A retrospective surveillance study has shown that the strain had been circulating in the Grampian region since 1999 and comprised about 1% of all MRSA cases, although only five other cases of infection in children were observed over this period. ST5-MRSA-IV isolates carrying the enterotoxin D gene but not the ETA gene were also isolated in Grampian and, more rarely, from elsewhere in Scotland. Not all Scottish diagnostic laboratories submit all new isolates to the Reference Laboratory so the full distribution of the outbreak strain is not known but the outbreak seems to have been caused by an exfoliative toxin-producing variant of a more widely distributed strain. The variation in PFGE pattern leaves open the possibility that the ETA-positive strain could have been generated from the ancestral ST5-MRSA-IV strain more than once.
CA-MRSA are often reported to be carrying the PVL gene and the strength of the association may be artificially enhanced by the virulence of isolates which produce this toxin. Patients outside the hospital system are almost certainly less likely to be swabbed than those in hospital and infections that are recurrent or difficult to treat are more likely to lead to laboratory investigations and detection of MRSA or MSSA. It may also be true that occurrence in a community hospital may make an outbreak more likely to be detected than occurrence in a group of patients with no apparent healthcare link. The detection of CA-MRSA outbreaks may therefore be biased towards those caused by toxin-producing strains and those with stronger healthcare links. Although none of the patients was seriously ill, production of the exfoliative toxin may have played some part in the clinical recognition of the spread of this strain. The outbreak stopped as suddenly as it had started, after almost 7 months and no further cases were observed under close observation in the following 6 months. Whether the standard infection control precautions contributed to this we cannot say. Widespread screening did not detect the outbreak strain in this or two other reports [Reference Bratu6, Reference Saiman7] casting doubt on this as a control strategy for outbreaks of CA-MRSA in the future.
Our cases had none of the usual predisposing factors associated with MRSA infection in children such as prematurity, low birth weight, underlying diseases, long duration of hospital stay or antibiotic exposure (except for two mothers) [Reference Campbell17, Reference de Almeida Silva18]. Perhaps because of this and the absence of the PVL gene, none was seriously ill, unlike in previous reports of PVL-positive MSSA and MRSA neonatal staphylococcal outbreaks [Reference Bratu6, Reference Saiman7]. Some of these outbreaks were also characterized by high colonization and transmission rates and spread to family members and the community. Only one of our cases (the first) seems likely to have acquired her infection in the community as it manifested 6 weeks after discharge. The majority of other strains identified in the surveillance study seem to have come from community cases (data not presented) so this strain can clearly spread in both hospital and community settings although it is not clear which came first.
This is the first ETA outbreak of CA-MRSA described, although several PVL-associated outbreaks, some from neonatal units, have been reported from around the world [Reference Chambers19–Reference Fridkin21]. Recently there have also been such reports from England [Reference Seetulsingh, Curtis and Kearns22–Reference David24]. None was caused by an exfoliative toxin-producing strain. There are several CA-MRSA strains, including some PVL positive, known to be circulating in Grampian and other parts of the UK. Presumably it is only a matter of time before other outbreaks are recognized although recent decline in community antibiotic use in the UK may have occurred just at the right time to slow the spread of these strains. MSSA isolates producing exfoliative toxins A and B are associated with Staphylococcal Scalded Skin Syndrome (SSSS) in neonatal units [Reference Ladhani25] and such isolates are occasionally referred to the Reference Laboratory. According to published accounts, these isolates belong to phage group II, unlike ST5-MRSA-IV (Scottish Reference Laboratory, unpublished data). Phage group II strains very rarely give rise to MRSA but it would be regrettable if avoidable failure to control isolates such as this outbreak strain led to the wide distribution of CA-MRSA capable of causing SSSS.
Nevertheless, this and the possible self-limiting outbreak described here, with relatively mild clinical manifestations, should not lull us into a false sense of security about the threat posed by CA-MRSA [Reference Gould26]. To this end interim guidelines on its control have been produced.
ACKNOWLEDGEMENTS
The authors thank all staff at the MRSA Reference Laboratory and the Diagnostic Laboratory, Foresterhill.
DECLARATION OF INTEREST
None.





