Introduction
Ruminants, in particular cattle, have been identified as a major reservoir of Shiga toxin-producing Escherichia coli O157:H7 (STEC O157) and related non-O157 STEC serogroups (e.g. O26, O45, O103, O111, O121 and O145) [Reference Moxley and Acuff1], which are recognised as important zoonotic pathogens worldwide [Reference Johnson, Thorpe and Sears2]. STEC can cause serious human illnesses such as haemorrhagic colitis and the potentially life-threatening haemolytic uraemic syndrome [Reference Gould3]. Frequent sources of STEC infection in humans are STEC-shedding animals or humans, faecally contaminated drinking and recreational water, or faecally contaminated food (reviewed in [Reference Karmali, Gannon and Sargeant4]).
Undercooked meat and meat products of bovine origin, contaminated with faecal material at slaughter, have been identified frequently as sources of foodborne STEC outbreaks in other countries [Reference Bell5–Reference Kassenborg7]. Although good hygiene practices on meat processing plants limit faecal contamination of carcasses at slaughter, hides have been identified as a major source of carcass contamination during processing of cattle [Reference Barkocy-Gallagher8–Reference Nou10], highlighting the importance of minimising faecal contamination of hides prior to slaughter. STEC-infected cattle shed the pathogen within their faeces that can then contaminate hides of other animals [Reference Arthur11–Reference Fegan13] as well as the environment during transportation and lairage (e.g. walls of transport trucks [Reference Cuesta Alonso, Gilliland and Krehbiel14] and lairage pen floors [Reference Childs12, Reference Small15]).
Studies in other countries have investigated the impact of transportation and lairage on faecal shedding of E. coli O157 [Reference Minihan16] and hide contamination with E. coli O157 [Reference Stanford17] in adult slaughter cattle, but no equivalent data are available for E. coli O157 and O26 in very young calves. This study focused on E. coli O26 and O157 because they are the two most common serogroups associated with human STEC disease in New Zealand (66% of all 2016 notifications where an isolate was obtained) [18] and because of the well-established fermentation characteristics/available media (e.g. CT-SMAC and CT-RMAC) for selective/differential culture of STEC O157 and O26, respectively. Other STEC such as O45, O103, O111, O121 and O145 are rarely associated with human infection, or live cattle in New Zealand, and selective media are unavailable for their culture.
In 2015, over 2.1 million very young calves were slaughtered in New Zealand [19] with most of the meat being exported. Calving on New Zealand dairy farms is timed to coincide with rapid forage growth in spring. This seasonal calving results in ‘surplus’ calves being slaughtered between July and October that are not reared as replacement stock or fattened for the beef market. This study was designed to determine the prevalence of faecal shedding of both toxigenic and non-toxigenic strains of E. coli O157 and O26, including STEC O157 and O26, in young calves at the time of slaughter and to investigate the effect of transportation and lairage on calf rectal carriage and contamination of hide and carcass through meat processing. Epidemiological data provided were used in the development of a risk management strategy for STEC in New Zealand.
Materials and methods
Pre-selection, pre-testing and classification of farms
During the 2010 calving season (July to September), a cohort study was conducted in the Waikato region, a large dairy farming area in the central North Island of New Zealand, following very young calves (4–7-day-old) from dairy farms to a regional slaughter plant. Dairy farms eligible for this study were selected from those that had participated in a nationwide cross-sectional study conducted at the same slaughter plant in the preceding year, and had calves that tested positive for E. coli O157 and O26 in faeces [Reference Jaros20]. Eight dairy farms (A1, A2, A3, B1, B2, C1, C2 and C3) from three different locations (A, B, C; 52–71 km distance between each location) in the Waikato region were pre-selected for this study. Pre-testing of dairy farms was completed within 4 weeks and overlapped with the 3-week study period (one study day per week), enabling the study to be completed within the calf processing season of the slaughter plant (Table 1).
Table 1. Study scheme with pre-testing of eight dairy farms in three locations in the Waikato region in the North Island of New Zealand

For pre-testing, all pre-selected farms from the same location were visited twice within a period of 2 weeks at the beginning of calving (i) to confirm E. coli O157 and O26 were present in calves, and then (ii) to estimate the prevalence of both serogroups in calves on each farm immediately prior to transporting calves for slaughter. On each visit, recto-anal mucosal swab (RAMS) samples from up to 10 calves per farm were collected, sampling only animals selected for slaughter. Each RAMS sample was enriched and screened for the presence of E. coli O157 and O26 by real-time PCR to assess on-farm prevalence of both serogroups (proportion of real-time PCR-positive calves).
Based on combined prevalence of E. coli O157 and O26 from both pre-tests on each farm, the pair of farms in each location (except B) that were most significantly different from each other were included in the study. Each farm was then classified as either a low-, medium- or high-prevalence farm for each serogroup.
Animal selection and description of transportation and lairage
On each study day, between 8 and 11 calves (numbers depending on availability) on both farms were selected and spray-marked before being picked up by a regional livestock transporter and transported collectively to the regional slaughter plant. Regional livestock transporters often pick up >100 calves per single truck from multiple (10–20) local dairy farms for delivery to a meat plant. On study day 1, calves from both farms were transported in the same crate pen on the transporter and not mixed with the calves from other farms. On study day 2, calves from both farms were transported in two separate crate pens and not mixed with other calves; while on study day 3, calves from both farms were transported in different crate pens but mixed with other calves. The duration of transportation from the farms to the slaughter plant was <2 h.
On arrival at the slaughter plant, the study animals from both farms were kept in the same holding pen for lairage and not mingled with other calves until being slaughtered early the following day. During lairage (<22 h), the study animals were able to have direct contact with neighbouring calves through gaps between the pen's rails and a shared water trough. The animals were not fed during lairage.
Sample and data collection
Multiple samples were collected from each calf as the cohort of study animals was followed on each study day from their farm of origin to the slaughter plant and along the processing chain.
Before slaughter, two RAMS samples were collected from each calf by swabbing the recto-anal junction with firm pressure using sterile cotton-tipped swabs (Transystem®, Copan, Italy), placed in Amies transport medium provided in the swab tubes and kept refrigerated until processed within 24–48 h of collection. The first RAMS was taken at the farm before transportation (‘on-farm’ sample) and the second on plant at the end of lairage immediately prior to slaughter (‘on-plant’ sample).
Post-slaughter, three sponge swab samples were aseptically collected from each carcass, using sponges pre-moistened with 10 ml of maximum recovery diluent (MRD, Fort Richard Laboratories, New Zealand), which were sealed in Whirlpak™ bags (Nasco, Fort Atkinson, WI, USA). The first sponge sample was taken from the hide near the opening Y-cut area before hide removal (‘hide’ sample); the second sample was taken from the opening Y-cut area and anal cavity on the left-hand side of the carcass post-evisceration but before the intervention was applied (‘pre-intervention’ sample), and the third sample from the opening Y-cut area and anal cavity on the right half of the carcass post-intervention (‘post-intervention’ sample) when entering the chiller. A hot water carcass wash (82 °C) was used as an antimicrobial intervention on the slaughter plant. Applying a left and right half of carcass sampling assumed that (i) both halves of the carcass were equally likely to be exposed to any possible contamination, and (ii) that variability of individual sampling between the left and right half of the carcass was negligible. All carcass swab samples were kept refrigerated until processed in the laboratory within 24 h of collection. Sex, breed, ear tag number and carcass weight were recorded for each calf.
Sample processing
Enrichment and direct culture plating of samples
Each RAMS was transferred into 20 ml of 100% tryptic soy broth (TSB) (Bacto™, Becton, USA), vortexed for 10 s, and 50 µl directly plated onto cefixime-tellurite sorbitol MacConkey agar (CT-SMAC, Fort Richard Laboratories) and cefixime-tellurite rhamnose MacConkey agar (CT-RMAC, Fort Richard Laboratories) used as selective culture media for E. coli O157 and O26, respectively. Direct culture plates were incubated at 37 °C for 18–24 h, while TSB broths were incubated at 25 °C for 2 h, followed by 42 °C for 6 h. For carcass samples, 90 ml of MRD (0.1% peptone, 0.85% sodium chloride, Fort Richard Laboratories) was added to each sponge swab sample and stomached for 2 min. One millilitre was taken immediately for a 10-fold serial dilution of each sample, which was plated onto CT-SMAC and CT-RMAC selective media and incubated at 35 °C for 24 h [21]. A 35 ml aliquot was stored at 4 °C for later analysis and the remaining volume (approximately 54 ml) was used for enrichment with the addition of 220 ml of 118% TSB and incubated at 25 °C for 2 h, followed by 42 °C for 6 h.
Detection of E. coli O157 and O26 serogroups
Each RAMS-enriched TSB broth was screened for the presence of E. coli O157 and O26 using an automated real-time thermocycler (Rotor Gene 6200HRM, Corbett Research, Australia). Genomic DNA was extracted from 1 ml of enriched TSB broth with 2% Chelex® beads solution (Bio-Rad, USA) and used in two separate PCR assays to detect genes encoding for serogroup-specific O-antigens of E. coli O157 and O26 as previously described [Reference Jaros20]. Similarly, for each carcass swab sample, genomic DNA was extracted from 1 ml of enriched TSB broth with PrepMan®Ultra (Applied Biosystems, USA) according to the manufacturer's instructions and screened for the presence of E. coli O157 and O26 using an automated real-time thermocycler (ABI TaqMan® 7300, USA) and probes to detect the rfbEO157 and wzx O26 genes of E. coli O157 and O26 [Reference Perelle22]. Each DNA sample was used in two single TaqMan® real-time PCR assays. The 20 µl TaqMan® real-time PCR reaction volume contained 0.4 µM of each primer (rfbEO157 or wzx O26), 0.2 µM of probe (FAM-labelled probes (ABI, Auckland, New Zealand)), 1× internal control (IC) DNA, 1× IC primer and probe mix (ABI), 3 mM of MgCl2 (Qiagen, Auckland, New Zealand), 2 µl of DNA and 10 µl of 2× TaqMan® master mix (ABI). The amplification programme included a standard cycling protocol: 50 °C 2 min, 95 °C 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s and annealing/amplification at 60 °C for 1 min. Fluorescence of the PCR product was measured at the end of the annealing step in channel A (520 nm).
All genomic DNA extractions were analysed immediately and then stored at −20 °C. Samples collected during field work were processed and analysed in two different laboratories with two different screening methods: real-time PCR for RAMS samples and TaqMan® for carcass swab samples. Hence, to ensure concordance between both methods, genomic DNA extracted from carcass swab samples was reanalysed with the real-time PCR applied on RAMS samples for confirmation of TaqMan® results, in order to show the results were comparable. The real-time PCR results were used for data analysis.
Culture isolation of E. coli
Real-time PCR- and TaqMan®-positive samples, including 10% randomly selected TaqMan®-negative samples, were subjected to culture confirmation. Direct culture plates of real-time PCR-positive samples were scanned for presumptive E. coli O157 and O26 colonies. For RAMS samples, up to 10 non-fermenting and 10 fermenting colonies per plate were tested with serogroup-specific latex agglutination kits and confirmed with real-time PCR as described previously [Reference Jaros20]. For the TaqMan®-positive samples, up to three non-fermenting and three fermenting colonies per direct culture plate were transferred to tryptic soy agar plates (Fort Richard Laboratories), incubated overnight at 37 °C. E. coli O157 and O26 presumptive isolates were identified using the same serogroup-specific latex agglutination. Confirmed isolates were preserved in glycerol broth (nutrient broth with 15% glycerol) and stored at −80 °C for further molecular analysis. To detect false-negative samples, colonies on direct culture plates of O157-negative real-time PCR samples were subjected to latex agglutination. This additional testing was undertaken for O157-negative real-time PCR samples only.
If E. coli O157 and O26 isolates could not be recovered from direct culture plates of real-time PCR- or TaqMan®-positive samples, immunomagnetic separation was applied to enriched TSB broths with inoculation onto CT-SMAC and CT-RMAC plates. Again, up to 10 non-fermenting and 10 fermenting presumptive E. coli O157 and O26 colonies per plate were tested with serogroup-specific latex agglutination kits and confirmed with real-time PCR as described previously [Reference Jaros20].
Allelic profiling and pulsed-field gel electrophoresis of isolates
Stored E. coli O157 and O26 isolates were resuscitated on Columbia horse blood agar (Fort Richard Laboratories). Genomic DNA was extracted from five colonies using 2% Chelex beads solution and analysed in two PCR assays to detect the presence of STEC virulence genes encoding for Shiga toxin 1 (stx1), Shiga toxin 2 (stx2), enterohaemolysin (ehxA) and intimin (eae), and stx2c (to differentiate from stx2a-positive O157:H7), as described previously [Reference Jaros23]. Each confirmed E. coli O157 and O26 isolate was subtyped using pulsed-field gel electrophoresis (PFGE; restriction enzyme XbaI) according to the standardised laboratory protocol published by PulseNet International [24].
Molecular analysis of isolates
BioNumerics software (version 6.6) [25] was used to analyse and compare PFGE profiles of E. coli O157 and O26 isolates, and to create a dendrogram applying unweighted pair group method with arithmetic mean cluster analysis using the Dice similarity coefficient, with a band-matching tolerance of 1%. Clusters of PFGE profiles were established with cut-off levels of 97.3% and 97.7% similarity for E. coli O157 and O26, respectively, and cluster numbers assigned. The assigned cluster numbers were also used to analyse the diversity of PFGE profiles by calculating the Simpson's diversity index (1–D) [Reference Simpson26] including 95% confidence intervals, using PAST software [Reference Hammer, Harper and Ryan27]. The Simpson's index ranges from 0 to 1, with values closer to 1 indicating a higher diversity of PFGE profiles.
Farm classifications and statistical analysis
Pre-study on-farm prevalence was calculated using samples from both pre-testing occasions and is defined as the percentage of the number of real-time PCR-positive animals divided by the total number of animals sampled on the farm. Similarly on study day, on-farm prevalence was calculated using the percentage of real-time PCR-positive animals divided by the number of animals sampled on the farm. The classification criteria for the prevalence were: ‘low’ if the prevalence value was ⩽30%, ‘medium’ if >30% and ⩽70% and ‘high’ if >70%.
R software (version 2.15.2) [28] was used for all statistical analyses. Multivariable logistic regression models, with and without random effects, were used to identify the risk factors associated with a calf being real-time PCR-positive for either serogroup of E. coli at slaughter (on-plant). Real-time PCR results were used for this analysis because of their higher prevalence and statistical power compared to culture results. The explanatory variables considered were: calf being real-time PCR-positive on-farm and the pre-study testing prevalence of E. coli (O157 or O26) on-farm. The multivariate models were generated and assessed for their significance and goodness-of-fit as described previously [Reference Jaros29].
Results
Pre-testing of farms prior to study days
RAMS from a total of 144 calves on eight selected dairy farms were collected for pre-testing to determine the prevalence prior to study days. All RAMS were screened by real-time PCR for the presence of E. coli O157 and O26 (STEC and non-STEC); the results are summarised in Table 2. Pairs of farms within each location that displayed the greatest contrast in the prevalence of E. coli O157 and O26 were selected for follow-up; hence, farms A1 and C1 were excluded from the remainder of the study.
Table 2. Recto-anal mucosal swab samples collected from 144 calves on eight pre-selected dairy farms in the North Island of New Zealand on two farm visits (two pre-tests) were screened for the presence of Escherichia coli O157 and O26 (STEC and non-STEC) by real-time PCR
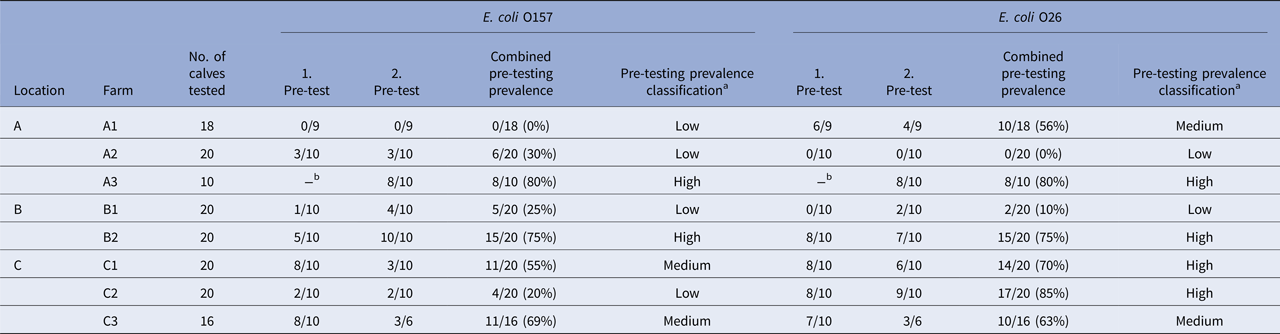
Results of pre-tests (real-time PCR-positive/samples tested), combined pre-testing prevalence from both pre-tests (%), and pre-testing prevalence classification of farms per serogroup, stratified by location and farm.
aClassification criteria for combined prevalence values: ‘low’ if prevalence ⩽30%, ‘medium’ if >30% and ⩽70% and ‘high’ if >70%.
bNo calves available.
Study animals and samples collected
A total of 60 calves were followed as cohorts from six dairy farms to slaughter on 3 study days, collecting 60 on-farm, 60 on-plant, 59 hide, 58 pre-intervention and 47 post-intervention samples. Number of animals and samples collected per study day and farm are summarised in Supplementary Table S1.
Real-time PCR and culture isolation
All collected samples were tested by real-time PCR for the presence of toxigenic and non-toxigenic E. coli O157 and O26 and confirmed by culture isolation; the results are presented in Table 3. No significant change in real-time PCR-positive RAMS samples was observed on-plant after transportation and lairage. Almost all hide samples were real-time PCR-positive for E. coli O157 and O26, with fewer real-time PCR-positive samples pre- and post-intervention. No consistent patterns could be observed in the culture isolation of E. coli O157 and O26 (STEC and non-STEC) among RAMS and carcass samples. Detailed results of real-time PCR and culture isolation of E. coli O157 and O26 (STEC and non-STEC), stratified by study day and farm, are presented in Supplementary Table S2. On-farm prevalence of E. coli O157 and O26 on study day is summarised in Table 4.
Table 3. Results of real-time PCR and culture isolation of Escherichia coli O157 and O26 (STEC and non-STEC) from pre- and post-slaughter samples collected from 60 calves, stratified by sample type
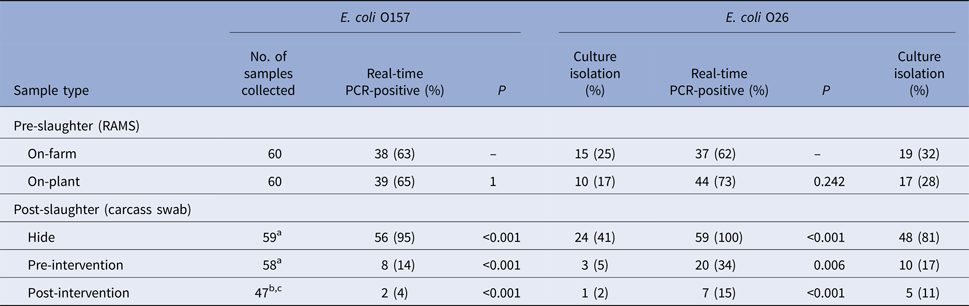
Loss of carcasses samples are explained in superscripts (a–c)
aCarcass missed on processing chain.
bCarcass condemned pre-intervention.
cIdentification tags lost in hot water wash (intervention).
Table 4. Results of on-farm prevalence (real-time PCR-positive/samples tested) and prevalence classification of farms on study day
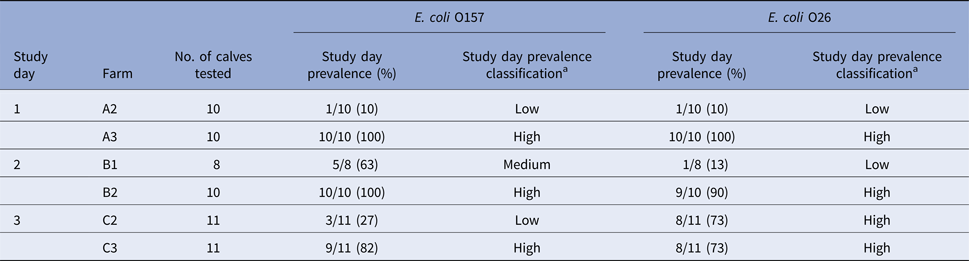
aClassification criteria for combined prevalence values: ‘low’ if prevalence ⩽30%, ‘medium’ if >30% and ⩽70% and ‘high’ if >70%.
Characterisation of confirmed isolates
In total, 56 E. coli O157 and 115 E. coli O26 isolates were retrieved and all were analysed by PCR for the presence of stx1, stx2, stx2c, eae and ehxA. Seventeen samples contained two or more different strain types of E. coli O157 or O26. Characteristics of all confirmed E. coli isolates, stratified by sample type and serogroups, are presented in Table 5. Twice as many E. coli isolates of serogroup O26 were recovered compared to O157; however, only 8.7% (10/115) of the E. coli O26 isolates were stx-positive. By contrast, 83.9% (47/56) of the E. coli O157 isolates were STEC. The highest diversity of different virulence types (stx1, stx2, stx2c and eae) of E. coli O157 and O26 was observed in the hide samples. The lowest numbers of toxigenic and non-toxigenic E. coli isolates were detected in post-intervention samples.
Table 5. Characteristics of Escherichia coli O157 (n = 56) and O26 (n = 115) isolates retrieved from recto-anal mucosal swab samples collected from 60 calves at pre-slaughter (on-farm, on-plant) and post-slaughter carcass swab samples (hide, pre- and post-intervention)
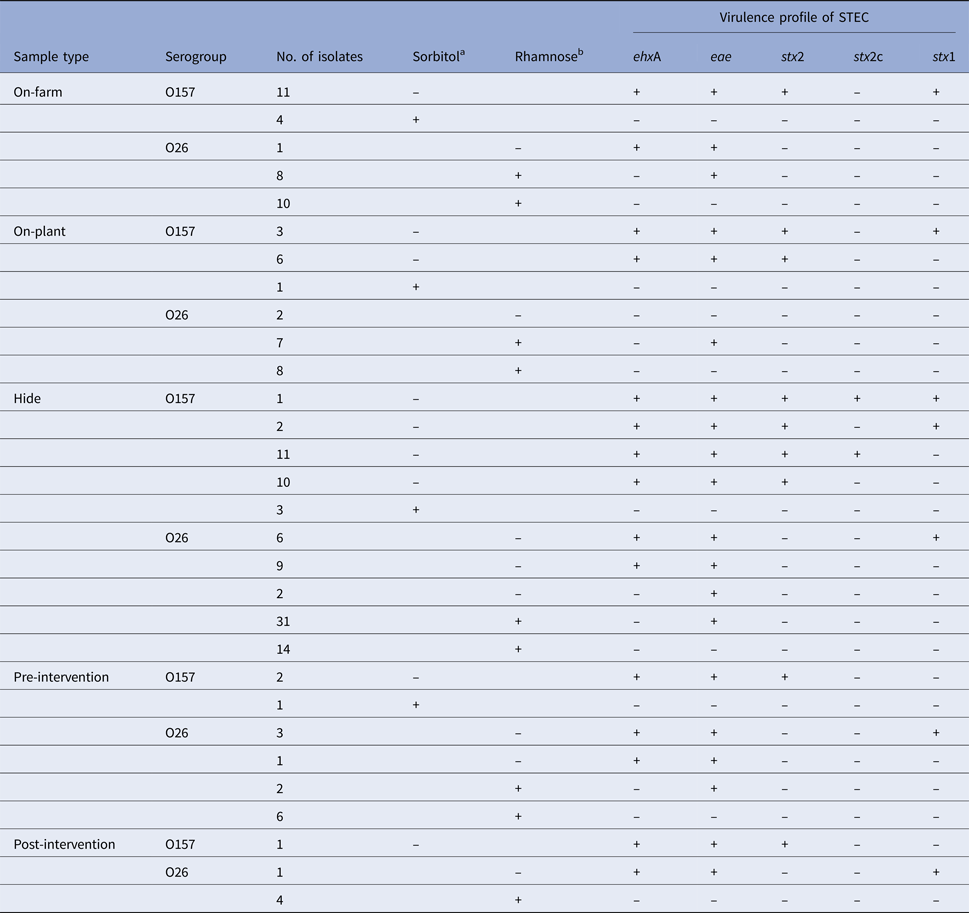
Isolates are stratified by sample type and serogroups. PCR methods were used to test for the presence (+)/absence (–) of virulence genes encoding for enterohaemolysin (ehxA), intimin (eae) and Shiga toxins (stx2, subtype stx2c, stx1).
aSorbitol-fermenting (+) or non-sorbitol-fermenting (–) on CT-SMAC culture plate.
bRhamnose-fermenting (+) or non-rhamnose-fermenting (–) on CT-RMAC culture plate.
Genotype diversity of isolates
Based on the cluster analysis of PFGE profiles, the diversity of all recovered E. coli O157 and O26 isolates (STEC and non-STEC) from different sample types is depicted in Figures 1 and 2, respectively. Cluster analysis of PFGE profiles of E. coli O157 and O26 isolates including virulence profiles is presented in Supplementary Figures S1 and S2, respectively. The genotyping results in Figure 1 provided no evidence that transmission of E. coli O157 infection (following ingestion, gut passage and colonisation) had occurred during transportation and lairage (i.e. there was no evidence of shared genotypes in faeces on-plant between co-transported calves from different farms). On post-slaughter hides, however, a high level of cross-contamination with genotypes (pale grey cells), which were not recovered from calves’ RAMS on-farm or on-plant, was observed.
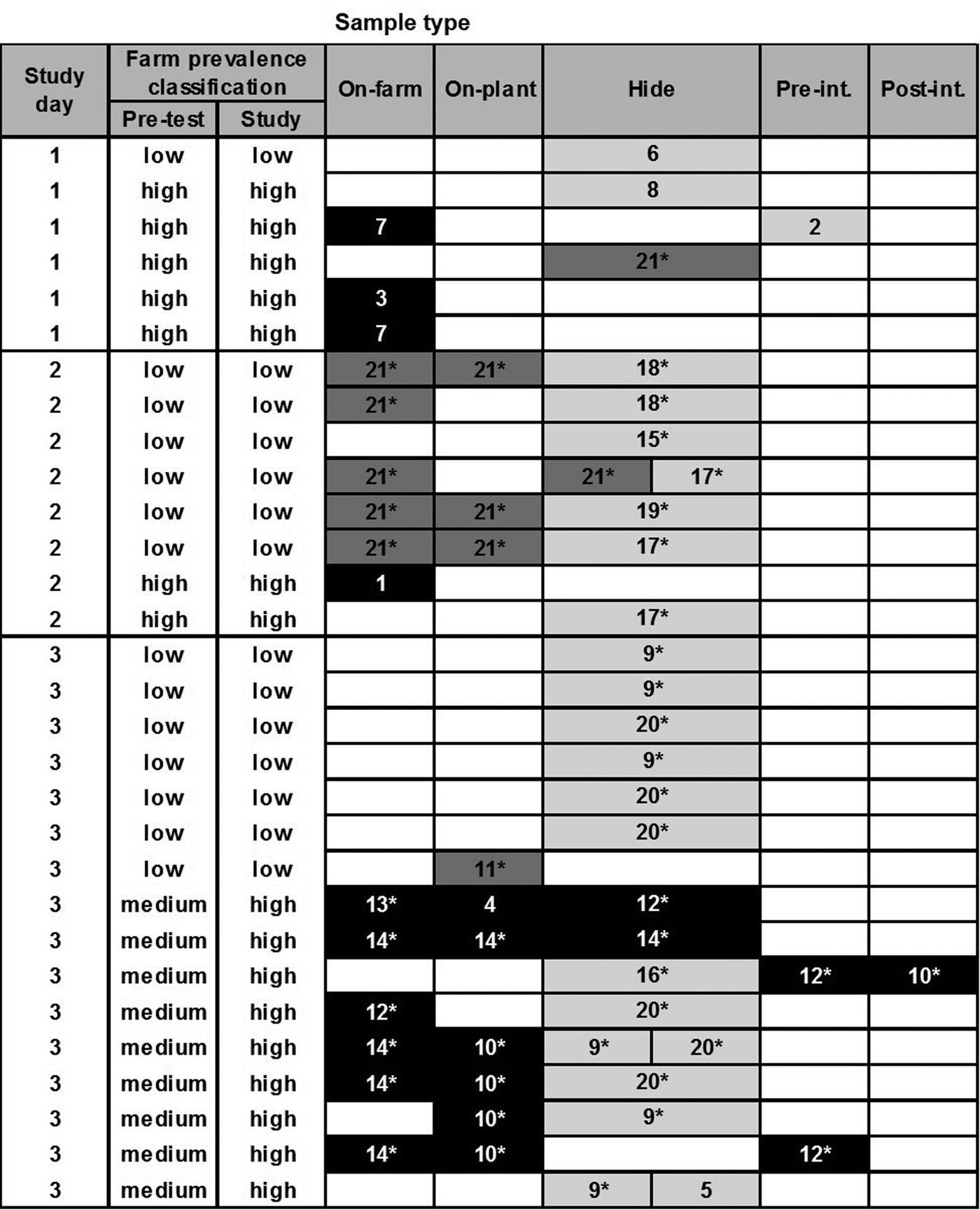
Fig. 1. Diversity of PFGE profiles of Escherichia coli O157 isolates (n = 56) recovered from different samples collected from 60 calves at pre-slaughter (RAMS: on-farm and on-plant) and post-slaughter (swab: hide, pre- and post-intervention). Only samples from animals with one or more recovered isolates are shown (each row), with split cells representing two characteristically different PFGE profiles of isolates, coloured by farm of origin. Black cells represent PFGE profiles prevalent on high-prevalence farms on study day, dark grey on low-prevalence farms and pale grey on neither low- or high-prevalence farms included in the study and therefore likely of different origin. Numbers in cells represent assigned PFGE cluster numbers and * identifies STEC isolates (see Supplementary Figure S1).
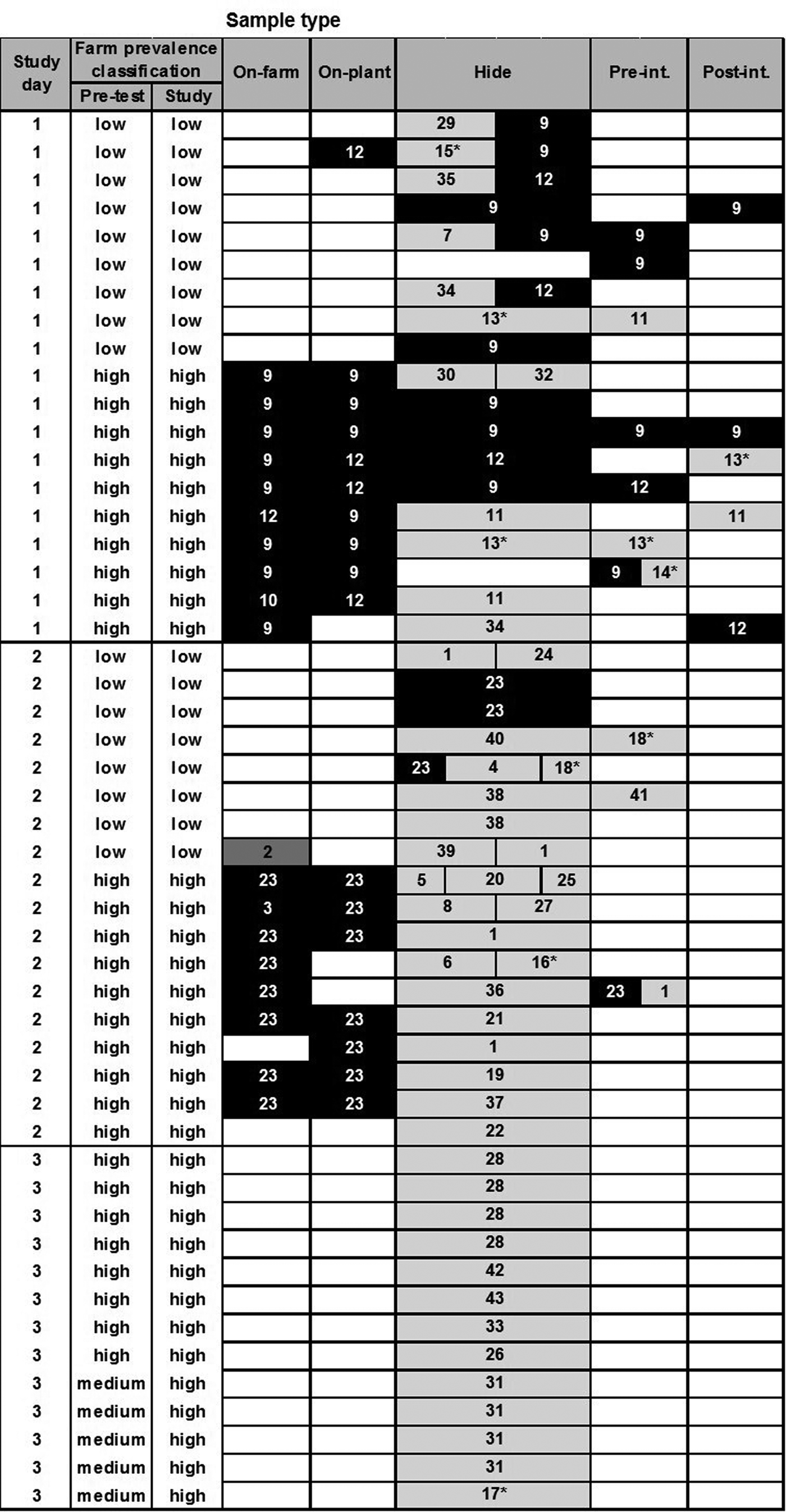
Fig. 2. Diversity of PFGE profiles of Escherichia coli O26 isolates (n = 115) recovered from different samples collected from 60 calves at pre-slaughter (RAMS: on-farm and on-plant) and post-slaughter (swabs: hide, pre- and post-intervention). Only samples from animals with one or more recovered isolates are shown (each row), with split cells representing up to three characteristically different PFGE profiles of isolates, coloured by farm of origin. Black cells represent PFGE profiles prevalent on high-prevalence farms on study day, dark grey on low-prevalence farms and pale grey on neither low- or high-prevalence farms included in the study and therefore likely of different origin. Numbers in cells represent assigned PFGE cluster numbers and * identifies STEC isolates (see Supplementary Figure S2).
For E. coli O26, there was some evidence of transmission between calves from different farms during transport and lairage (Fig. 2); one calf from a low-prevalence farm was faecally positive on-plant with PFGE type 12, a genotype also isolated from co-transported calves from the high-prevalence farm on-farm and on-plant. There was stronger evidence of hide and carcass cross-contamination pre-intervention from high- to low-prevalence animals on study days 1 and 2, and evidence of hide contamination with multiple genotypes not identified in animals on the study farms. Despite this, there was no evidence of a difference in residual contamination on the carcasses of calves from high- and low-prevalence farms post-intervention.
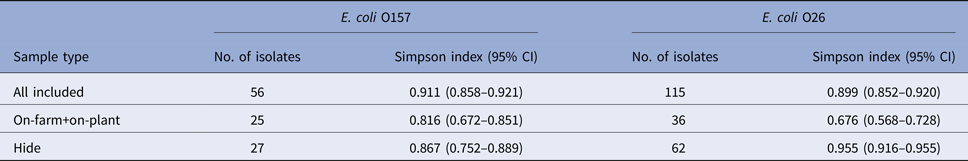
To demonstrate the diversity of E. coli O157 and O26 isolates from specific sample types, Simpson's index (1–D) values were calculated using cluster numbers from the PFGE profile analysis of isolates (Table 6). A high diversity was observed for both serogroups when analysing all isolates collectively; however, after stratification by sample type, the point estimate of the diversity index was higher at the hide stage for both serogroups. For O26, the 95% confidence intervals for the diversity estimates between pre-slaughter (on-farm and on-plant isolates combined) and hide samples did not overlap, indicating a statistically significant difference (P < 0.05).
Table 6. Simpson's index (1–D) values with 95% confidence intervals (CI) showing the diversity of PFGE profiles of Escherichia coli O157 (n = 56) and O26 isolates (n = 115) recovered from pre-slaughter recto-anal mucosal swab samples (on-farm, on-plant) and post-slaughter carcass swab samples (presenting hide only) from 60 calves
Risk factors for faecal carriage at slaughter
On univariable analysis, a calf being real-time PCR-positive for E. coli O157 in faeces on-plant was positively associated with the calf being real-time PCR-positive on-farm and the E. coli O157 prevalence on the farm of origin at pre-testing. When both variables were included in a multivariable model, only the calf being positive on-farm was significantly associated with being positive on-plant (Table 7). Similarly to E. coli O157, a calf being real-time PCR-positive for E. coli O26 in faeces on-plant was also associated with the calf being real-time PCR-positive on-farm and on-farm prevalence during pre-study testing on univariable analysis (Table 7). When both variables were included in a multivariable model, only on-farm prevalence was significantly associated with being positive on-plant.
Table 7. Models and results of multivariate logistic regression analysis for a calf being real-time PCR-positive (Escherichia coli O157 or O26) in faeces at slaughter (‘on-plant’ after transportation and lairage)
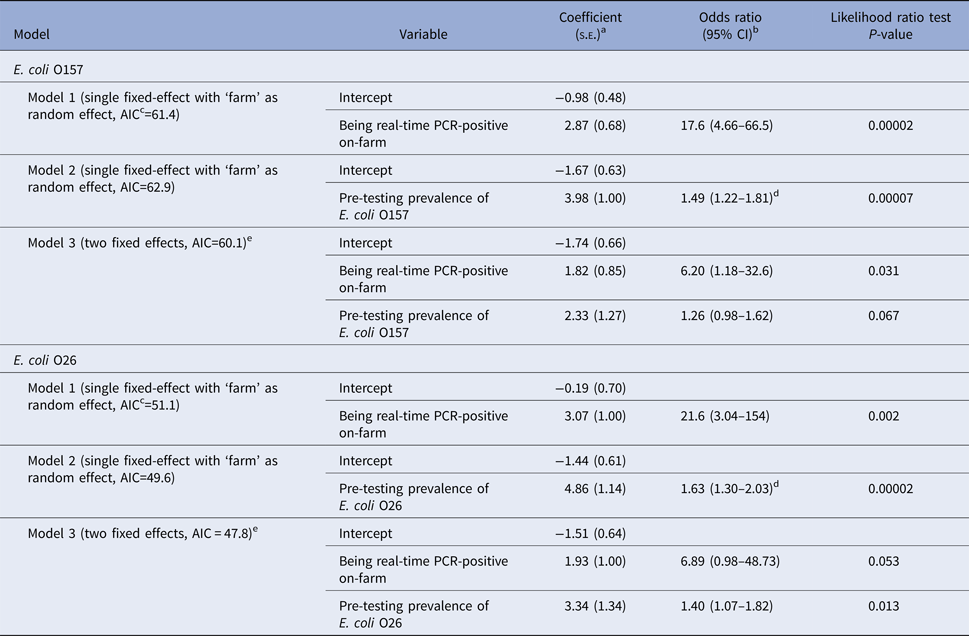
Models presented separately for E. coli O157 and E. coli O26 (60 observations per serogroup).
as.e.: standard error.
bCI: 95% confidence interval.
cAIC: Akaike Information Criteria, a measure of the relative quality of each model, with lower values indicating better quality.
dThe odds ratio of this continuous variable refers to an increase in the prevalence of 0.1 (10%) on the farm.
eModel with and without random effect identical due to unique covariate pattern for each farm.
Discussion
To the best of our knowledge, this is the first study to investigate the effect of short-haul transportation and lairage on faecal shedding and carcass contamination of E. coli O157 and O26 (STEC and non-STEC) in very young calves. This study has provided important epidemiological data to highlight potential critical control points in the contamination of veal carcasses with E. coli O157 and O26 after transportation and lairage under New Zealand conditions.
Effect of transportation and lairage on faecal shedding
A slight increase in real-time PCR prevalence of faecal shedding of E. coli O157 or O26 was observed in calves after transportation and lairage at the slaughter plant (<24 h); however, this was not significant and not supported by culture isolation. The presence of one calf at slaughter that was positive for an E. coli O26 PFGE genotype 12 that was otherwise only recovered from co-transported calves from a high-prevalence farm, may indicate transmission during transport and lairage. It is possible that some calves had ingested E. coli during transportation and lairage but either the pathogen did not reach the recto-anal junction to be detected at the time of sample collection on-plant, or E. coli was shed in concentrations below the detection limits of the culture methods used. Older calves exposed to STEC O157 shed the pathogen within 1 day/24 h in studies where 3-month-old bull calves (natural transmission) [Reference Lahti30], calves of body weight 97.8 ± 16.6 (s.d.) kg (natural transmission via exposure to inoculated house flies) [Reference Ahmad, Nagaraja and Zurek31] or 6–9-month-old steers (oral inoculation of E. coli) were examined [Reference McGee32].
Transportation has been identified as a potential stressor likely to induce faecal shedding of Salmonella spp. in feeder calves [Reference Corrier, Purdy and Deloach33] and adult feedlot cattle [Reference Barham34], but could not be confirmed for E. coli O157 in the study by Barham et al. [Reference Barham34]. It appears that transportation of only a few hours has no or very little impact on the prevalence of faecal shedding of E. coli O157 in cattle. Minihan et al. [Reference Minihan16] did not observe any increased prevalence of E. coli O157 in faeces from adult feedlot cattle after 1.5 and 6 h transportation to slaughter plants in Ireland. Similarly, Bach et al. [Reference Bach35] reported no increased faecal shedding of STEC O157 in steer calves (220 ± 37 kg) after relocation to a feedlot in 3 h (short-haul transportation, with or without transport preconditioning of animals), in contrast to a 15 h transportation (long-haul, without preconditioning). Although no faecal samples were collected from calves on arrival at the slaughter plant in our study, previous studies suggest that short-haul transportation (<2 h) is less likely to induce or increase the faecal shedding of E. coli in very young calves.
‘Being positive on-farm’ and ‘on-farm prevalence’ 1–2 weeks prior to study days were identified as risk factors for test calves being real-time PCR-positive in faeces after transportation and lairage on-plant (Table 7). These findings indicated that the real-time PCR status of an animal on-plant was strongly associated with E. coli prevalence at the animal/farm level. Housing/farm management factors and age of calves have been identified as risk factors for increased prevalence of STEC O157 on dairy farms in the previous studies [Reference Garber36, Reference Nielsen37]. Cobbold and Desmarchelier [Reference Cobbold and Desmarchelier38] reported that higher prevalence of STEC in dairy calves was associated with group penning of animals, with pen floors and calf hides as important means of horizontal STEC transmission between calves. These observations support our findings as it is general practice on New Zealand dairy farms to keep very young calves in groups in barns, pointing to husbandry as a critical control point to reduce the prevalence of STEC at the farm and animal level. Hence, faecal shedding of E. coli in very young calves on-plant is less likely to be affected by transportation and lairage under New Zealand conditions, but more likely to be associated with farm-related factors that result in exposure and colonisation by STEC.
Effect of transportation and lairage on hide contamination
Almost every hide sample was positive by real-time PCR or culture. As no comparable hide samples were collected from calves before and after transportation, the impact of transportation and/or lairage on hide contamination cannot be evaluated. However, there was an indication of post-farm contamination of hides during transport and lairage as some PFGE profiles of E. coli not detected in the on-farm samples appeared in calves from both high- and low-prevalence farms on the same study day; e.g. PFGE type 17 on study day 2 and PFGE types 9 and 20 on study day 3 for E. coli O157; and similarly for E. coli O26 with PFGE types 11, 13 and 34 on study day 1 and PFGE type 1 on study day 2. Of note is that stx2c-positive E. coli O157 (PFGE types 17–20) were only associated with post-farm contamination of hides, and were neither isolated from calves before transport, or carcass swabs pre- or post-intervention highlighting good hygienic dressing practices at the slaughter plant. Previous studies have identified the use of commercial transportation as a (borderline) significant risk factor for increased E. coli O157 cross-contamination of cattle hides at slaughter [Reference Mather39], and holding animals in E. coli O157-positive or faecally contaminated lairage pens have also been described as a higher risk of hide contamination in finished beef cattle [Reference Dewell40]. Similarly, Arthur et al. [Reference Arthur41] reported that the transfer of STEC O157 onto cattle hides in the lairage area accounted for a larger proportion of hide and carcass contamination than the initial level of STEC O157 found on hides when cattle left the feedlot. Hence, a further study of similar design, which includes the collection of hide samples from calves before and after transportation, would be required to determine if transportation or lairage has the greater impact on the hide contamination of calves at slaughter under New Zealand conditions, or whether hide contamination occurs mainly on farms.
Based on PFGE profiles of E. coli isolates, cross-contamination of hides with E. coli O157 and O26 from calves originating from high- to low-prevalence farms was observed on study days (particularly for E. coli O26), in addition to a very high level of cross-contamination with E. coli isolates from different origins. Although no environmental samples were collected for the purpose of comparison, the cross-contamination of hides with genotypes different to those isolated from faecal samples of calves from high- and low-prevalence farms was most likely attributed to residual faecal contamination of vehicles and lairage yards, and mixing of study animals with calves from other farms during transportation (study day 3). Hide contamination via the environment was described by Childs et al. [Reference Childs12] when STEC O157 isolates from transport trailer side walls and pen side rails on-plant matched genotypes of isolates found on the hides of feedlot cattle at slaughter. Arthur et al. [Reference Arthur11] reported that only 29% of STEC O157 isolates detected on the hides of feedlot cattle at post-slaughter matched genotypes collected from the hides before transportation. Combined with our observations, this indicates that a large proportion of hide cross-contamination in cattle (adult and calves) maybe due to residual faecal contamination in the environment.
In addition to transportation and lairage, there are also animal-related factors affecting the prevalence of hide contamination in cattle. For example, the presence of ‘supershedders’ in the pens of feedlot cattle can significantly increase transmission [Reference Spencer42] and the prevalence of hide contamination [Reference Arthur43]. Similarly, Jacob et al. [Reference Jacob, Renter and Nagaraja44] found a significant correlation between hide prevalence within truckloads (a cohort of adult cattle transported and lairaged together) and the presence of high shedders (>5 × 104 CFU/g faeces) in truckloads, indicating that the levels of faecal E. coli shed during transportation and lairage also have an impact on the prevalence of hide contamination among slaughter cattle. However, the role of young calf ‘supershedders’ in post-farm STEC transmission has not been established.
Carcass contamination at pre- and post-intervention
This study suggests contamination of carcasses with E. coli O26 between calves from high- and low-prevalence farms, and possibly from the environment and other animals. Slaughter practices have a large effect on the microbiological contamination of red meat carcass [Reference Roberts45], with hides recognised as one of the main sources of carcass contamination with E. coli (and other pathogens of public health concern) during beef processing [Reference Barkocy-Gallagher8, Reference Arthur46–Reference Bell48]. Apart from the animal component, there are several plant-specific factors, which also have an effect on the extent of carcass contamination, such as plant design, processing speed, degree of good handling practices, and skills of abattoir personnel [Reference Hudson, Mead and Hinton49, Reference Biss and Hathaway50]. Hence, the carcass contamination results presented in this study are specific for the slaughter plant and may not be representative for other young veal processers across New Zealand.
Despite the evidence of cross-contamination of hides and carcasses pre-intervention, this was not evident for carcasses post-intervention, with only two out of nine real-time PCR-positive swab samples yielding STEC O157 or STEC O26 isolates at post-intervention. The overall prevalence of cross-contaminated hides and carcasses at pre- and post-intervention stages might have been higher as 15 out of 60 post-slaughter samples were not collected on study day 1 (loss of follow-up of carcasses). Nevertheless, this study suggests that the intervention used was effective in eliminating/reducing E. coli O26 and O157 contamination of carcasses, although the study was not designed to evaluate the efficacy of the hot water wash used at the slaughter plant. Extensive research in pre- and post-slaughter intervention strategies has been conducted and are still ongoing to develop novel methods to reduce the prevalence/level of STEC E. coli in/on live cattle and processed carcasses ([Reference Elramady51, Reference Koohmaraie52], cited in [Reference Sargeant53]). Some chemical and hot water washes are intervention methods currently approved for use on New Zealand veal slaughter plants.
Public health perspectives
In this study, we assume that O157:H7 isolates that are stx2-positive and stx2c-negative are stx2a-positive. This assumption is based on the previous work [Reference Jaros23] that analysed 403 STEC O157 isolates from humans and cattle across the North and South Island between 2008 and 2011. Using conventional PCR (generic stx2 primers and stx2c subtyping) and stx-encoding bacteriophage insertion (SBI) typing, the previous study showed that all isolates were either stx2c- or stx2a-positive or, on one occasion, positive for both virulence genes. The non-toxigenic O26 and O157 variants isolated in this study are likely to have limited food safety/public health significance. However, on some occasions, E. coli have been isolated from human clinical cases of disease where the stx gene is thought to have been lost [Reference Bielaszewska54]. The stx-negative eae-positive O26 are likely to be atypical EPEC (enteropathogenic E. coli), rarely associated with severe outbreaks of human diarrhoeal disease, and subsequent whole-genome sequencing of these isolates indicates that they are a separate phylogenetic lineage compared with STEC O26 [Reference Ogura55]. Similarly, whole-genome sequencing and preliminary characterisation of the sorbitol-fermenting stx-negative O157 have indicated that many lack virulence factors associated with human diarrhoeal disease and are very likely to be avirulent [Reference Iguchi56].
Conclusions
Although the number of selected farms and calves was relatively small, this study provides evidence that short-haul transportation and lairage under New Zealand conditions is not associated with increased faecal shedding of E. coli O157 and O26 in calves at slaughter, but associated with increased cross-contamination of hides with E. coli O26 between calves from high- and low-prevalence farms, and E. coli O26 and O157 strains from undetermined sources. The study did not demonstrate an increased prevalence of contaminated carcasses post-intervention, highlighting the need for good hygienic dressing practices and the impact of effective antimicrobial interventions during meat processing.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0950268818000973
Acknowledgements
The authors thank all participating dairy farmers and livestock transport companies, as well as the slaughter plant manager and staff for their great collaboration and participation in this study; the Food Microbiology and Safety Team from AgResearch (Ruakura, Hamilton, New Zealand) for collection and processing of post-slaughter samples; and laboratory staff from mEpiLab, Lynn Rogers, Julie Collins-Emerson, and Anne Midwinter for technical support and assistance. This work was supported by the Ministry for Primary Industries, Wellington, New Zealand, and the New Zealand Meat Industry Association.
Declaration of interest
None.
Ethical standards
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008. The study was approved by the Animal Ethics Committee of Massey University, Palmerston North, New Zealand; protocol number 10/44.












