INTRODUCTION
Escherichia coli is the most common bacterial species involved in urinary tract infection (UTI), which may lead to secondary life-threatening infections such as bacteraemia. Most infections are caused by extraintestinal pathogenic E. coli (ExPEC) clones present in the patient's own faecal flora. ExPEC clones belong mainly to E. coli phylogenetic groups B2 and D, and possess virulence factors that are uncommon in commensal or diarrhoeagenic clones [Reference Russo and Johnson1]. Although UTIs are typically endogenous infections, evidence has accumulated showing that external reservoirs such as sexual partners, children, siblings and pets may play an important role in the initial acquisition of ExPEC clones prior to infection [Reference Foxman2, Reference Manges, Johnson and Riley3]. A recent cross-sectional survey by Johnson et al. [Reference Johnson4] documented extensive sharing of faecal E. coli clones in multiple members of the same household. The authors hypothesized that clone sharing could reflect within-household transmission and that host-to-host transmission may contribute to the pathogenesis of UTI through dissemination of ExPEC clones from carriers to people living in close contact.
Investigating temporal shedding patterns of E. coli rather than diversity at a single sampling point has been recognized as the optimal way of detecting clone sharing within family households [Reference Johnson and Clabots5, Reference Johnson, Clabots and Kuskowski6]. The current knowledge of temporal E. coli shedding patterns in humans and pets is limited to few studies based on a single person [Reference Sears, Brownlee and Uchiyama7, Reference Caugant, Levin and Selander8] or household [Reference Johnson and Clabots5, Reference Johnson, Clabots and Kuskowski6, Reference Murray, Kuskowski and Johnson9, Reference Gordon, Bauer and Johnson10]. Most studies employed low-discriminatory typing methods such as serotyping [Reference Sears, Brownlee and Uchiyama7] or multilocus enzyme electrophoresis [Reference Caugant, Levin and Selander8, Reference Gordon, Bauer and Johnson10], and therefore were not able to elucidate entirely the actual diversity of the E. coli population in faeces.
We conducted a longitudinal study on 18 humans and 13 dogs living in eight households. Temporal changes in the shedding patterns of individual hosts and clone sharing within households were investigated by genotyping of 322 E. coli isolates obtained over a period of 6 months. Shedding patterns of presumptive ExPEC were inferred by examining the prevalence of phylotypes associated with UTI. In addition to random isolation, an antimicrobial selective culture method was used to enhance detection of low-prevalence clones shared within families.
MATERIALS AND METHODS
Recruitment of participants and sampling
Eight dog-owning families were recruited by a local advertisement at the University of Copenhagen. Details on age, gender and intra-family relations of the 31 participants (18 humans and 13 dogs) are described in Table 1. The two inclusion criteria for participation were ownership of at least one dog and availability to provide faecal swabs from all household members according to a defined sampling schedule. Samples were collected from June to December 2007 on days 1, 3, 11, 18, 25, 57, 92, 128, 155 and 183, and sent to our laboratory by ordinary mail. The procedure of sampling was explained to the participants by oral and written information. Human participants were asked to provide rectal swabs (BBL CultureSwab, Becton Dickinson, USA) or swabs dipped into faeces voided on a clean surface. All canine samples were collected following the latter procedure. Information on feeding habits of the dogs and on the household members' level of physical contact to their dogs was obtained by a specific questionnaire. The protocol of the study was approved by the Danish National Committee on Biomedical Research Ethics (H-KF-2007-0007).
Table 1. Description of participants from eight households and diversity of E. coli isolated non-selectively from each subject during 6 months

AFLP, Amplified fragment length polymorphism.
* Diversity index was calculated for each subject as number of AFLP profiles divided by number of isolates.
Bacterial isolation and identification
Each swab was streaked on MacConkey agar (Oxoid, UK) to obtain single colonies. After overnight incubation at 37°C, one presumptive E. coli colony was randomly selected, subcultured on blood agar and stored in brain heart infusion and glycerol at −80°C. In order to enhance detection of low-prevalence clones, swabs were also subjected to a selective direct plating method (DPM) [Reference Bartoloni11]. In brief, faecal swabs were uniformly streaked on MacConkey agar and the following antimicrobial discs (Oxoid) were applied onto the agar surface: amikacin (30 μg); cefotaxime (30 μg); ciprofloxacin (5 μg); ampicillin (10 μg); gentamicin (10 μg); tetracycline (30 μg) and sulphamethoxazole–trimethoprim (23·75/1·25 μg). The resistance patterns were read after overnight incubation at 37°C. Lactose-positive colonies growing in proximity of the discs, or within the inhibition zones determined by the discs, were selected if the same resistance phenotype was detected in another member of the household at the same sampling time.
E. coli was identified biochemically based on lactose fermentation, production of indole and failure to grow on citrate agar. The phylotypes of confirmed E. coli isolates were determined by multiplex PCR [Reference Clermont, Bonacorsi and Bingen12].
Detection of extended spectrum β-lactamase (ESBL) producers
E. coli isolates growing in proximity of cefotaxime discs were analysed for ESBL production by the double-disc diffusion method [13] using cefotaxime (30 μg) and cefotaxime combined with clavulanic acid (30/10 μg). PCR and sequence analysis were performed on putative ESBL producers for identification of CTX-M genes [Reference Hasman14].
Amplified fragment length polymorphism (AFLP)
AFLP was performed on all E. coli obtained by non-selective isolation as well as on selected isolates recovered by antimicrobial selective isolation, i.e. all isolates sharing the same antimicrobial resistance phenotype and E. coli phylotype within each household. The AFLP protocol used was modified from a previously developed method for typing of Salmonella spp. [Reference Torpdahl and Ahrens15]. Briefly, chromosomal DNA from isolates was digested by the enzymes BspDI and BglII followed by ligation of digests to specific oligonucleotide adaptors and PCR amplification of ligation products (for details see Supplementary material, available online). AFLP fragments were separated and detected on an ABI 377 automated sequencer (PerkinElmer, USA). One isolate (F4-1-tet) was included in all pre-run steps and included on all gels to allow evaluation of the reproducibility of the method. Isolates yielding weak or unclear bands were subjected to typing a second time. AFLP profiles were analysed with the GelCompar II software (Applied Maths, Belgium) using the Pearson product-moment correlation coefficient and clustering by the unweighted-pair group method with arithmetic averages (UPGMA). Band position tolerance and the optimization coefficient were set to 2·0% and 6·0%, respectively. The lowest similarity detected between identical samples run repeatedly was 85%. Consequently, AFLP fingerprints were considered as distinct when band patterns shared <85% similarity (cut-off). Visual inspection of electropherograms and epidemiological information were used to classify isolates close to the cut-off as distinct or indistinguishable.
Definitions
A clone was defined as one or more E. coli isolates belonging to one unique AFLP profile. According to the definition by Nowrouzian et al. [Reference Nowrouzian16], recurrent clones isolated on two or more occasions at least 3 weeks apart from the same subject were defined as resident and all other clones were considered transient. Shared clones were clones isolated from more than one subject. Clone sharing was defined as ‘within household’ when it involved subjects in the same household and defined as ‘across household’ when clones were shared between subjects in distinct families.
Data analysis
The level of clone sharing within households was compared to the level of clone sharing across households using the approach and definitions described by Johnson et al. [Reference Johnson4]. A diversity index (DI) was calculated as a measure of genotypic diversity in randomly isolated E. coli. The index was calculated for each subject as the number of clones divided by number of isolates (Table 1). To determine any differences between diversity in humans and dogs, the average DI was calculated for each host species.
Statistical analyses were performed solely on data retrieved from randomly chosen isolates obtained non-selectively. Mean values of diversity indices were compared using Student's t test and comparison of proportions was done using the χ2 test and Fisher's exact test. The threshold for statistical significance was established at P<0·05.
RESULTS
Occurrence of E. coli and ESBL producers
A total of 295 faecal swabs were received during the 6 months of sampling, resulting in a response rate of 97%. E. coli was isolated non-selectively from 264 swabs (89%). In addition, 113 E. coli isolates were obtained by DPM and 58 of these were selected based on phylotyping, leading to a total of 322 E. coli subjected to AFLP typing. ESBL producers were detected in two samples originating from the same dog (A4) and both isolates contained genes belonging to the CTX-M9 group.
Distribution of clones and phylotypes
A total of 154 distinct profiles (clones) were detected in the 322 isolates analysed by AFLP. Two isolates yielded consistently weak bands and were considered as non-typable. Phylotype A was most common in human clones (32%) whereas B1 was the most frequent in canine clones (42%) (Table 2). Of the 264 randomly obtained isolates, 136 distinct clones were detected. Sixty-four clones (47%) were found in dogs only, 61 (45%) in humans only and 11 (8%) occurred in both humans and dogs. Fourteen clones (10%) were shared across households and 12 clones (9%) were shared within households. Seven additional clones were detected by DPM, leading to a total of 19 clones shared within households (Table 3). Forty-one clones (27%) were detected in the same subject on multiple sampling times and 21 of them (13%) were defined as resident.
Table 2. Distribution of phylotypes in E. coli clones occurring specifically in humans and dogs
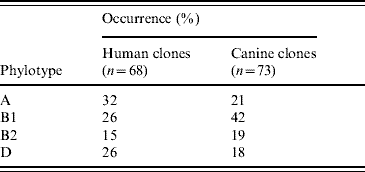
Table 3. Distribution of the 19 E. coli clones shared within six households

* Numbers refer to clonal types identified by amplified fragment length polymorphism and fields denoted by ‘–’ indicate that no clones were shared with other household members.
Diversity and shedding patterns of E. coli
The shedding patterns of randomly obtained E. coli isolates varied substantially in individuals (Table 1). The overall average DI was 0·66, indicating that about seven distinct clones occurred in 10 isolates. For individual participants the value ranged from 0·1 (10 identical isolates) to 1·0 (10 distinct isolates). The average DI was significantly higher in dogs (0·80) compared to humans (0·55) (P=0·01). One or two resident clones were detected in most humans (72%) and dogs (69%).
Clone sharing across and within households
The study population included 465 potential pairs sharing the same E. coli clone, including 51 within-household sharing pairs and 414 across-household sharing pairs. Based on randomly obtained isolates, clone sharing occurred significantly (P<0·001) more frequently in within-household pairs (12/51, 24%) than in across-household pairs (14/414, 3%). Human and canine members of four households shared the same clone at least once (Table 3). Sharing between humans was detected in three of the six households where more than one human was sampled. No clone sharing occurred in the two households (E and H) consisting only of one dog and its owner. Of the 19 clones shared within households, 11 belonged to phylotypes B2 (n=7) and D (n=4). The questionnaire data revealed that (i) all dogs were stroked daily by their owners, (ii) 85% of dogs licked their owner's faces regularly (⩾1/week), (iii) 54% of dogs stayed in their owner's beds or furniture regularly (⩾1/week), and (iv) 46% of dogs were fed table scraps regularly (⩾1/week). Several clones were shared between humans and dogs in family D (Table 3) despite stroking being the only reported physical human–dog contact. Only two persons (B1 and B2) practised hand-washing upon stroking their dogs. These persons did not share any clones with their dogs.
DISCUSSION
This is the first study to investigate E. coli shedding patterns in humans and dogs from several households over an extended period of time. Long-term carriage or intermittent shedding of one or two resident E. coli clones was demonstrated in most humans and dogs (~70%) by using a random isolation approach, which allows detection of mainly highly prevalent clones. This finding is consistent with previous experimental and observational studies demonstrating that individual humans and dogs are colonized by one or two resident E. coli clones at any given time [Reference Sears, Brownlee and Uchiyama7, Reference Sears17]. The reason why some E. coli clones persist in the intestine has been gradually elucidated. Early studies suggested that resident E. coli had special characters such as colicin resistance [Reference Hartl and Dykhuizen18] and later it was shown that genes encoding P fimbriae were enriched in resident E. coli clones [Reference Tullus19, Reference Wold20]. Recently, E. coli belonging to phylogenetic group B2 were found to be associated with persistence in the intestine [Reference Nowrouzian, Wold and Adlerberth21]. Our data based on phylotyping of random isolates support this notion since 15/21 detected resident clones (71%) belonged to group B2. We found that the overall distribution of phylotypes (Table 2) was somewhat similar for human and canine clones although human clones were slightly more likely to belong to groups B2 and D (41%) in comparison with canine clones (37%). Most of the canine clones (42%) belonged to group B1, a phylogenetic group highly associated with E. coli of animal origin [Reference Escobar-Paramo22].
Despite the occurrence of resident clones in most individuals, the composition of the E. coli population in human and dog faeces appeared to vary considerably over time. Except for two participants (B2 and F2), from which indistinguishable isolates were recovered throughout the study, most humans and dogs were found to shed multiple clones. This observation is exemplified by the high average DI (0·58) observed in humans, indicating that about six distinct clones were detected in 10 randomly selected isolates from the same individual. The overall diversity of E. coli clones in dogs was even higher as determined by comparison of the average DIs in the two hosts (P=0·01). This difference could be due to the exploratory behaviour typical of dogs, which results in higher exposure to external sources of E. coli. Indeed, most owners reported that their dogs regularly drank from puddles or consumed dead birds or faeces of cat or horse origin. It has been previously shown that human intestinal E. coli populations are less diverse over time than in animals such as cows and horses [Reference Anderson, Whitlock and Harwood23].
A high overall diversity was also observed by typing the 113 isolates recovered selectively by DPM. These isolates represented 53 cases of presumptive clone sharing within households but the results of phylotyping narrowed the cases of possible clone sharing down to 28. Further typing by AFLP showed that only seven cases of clone sharing were present. Thus, although each of the 53 cases represented two or three spatially and temporally related isolates with the same resistance phenotype, clone sharing was confirmed in only 13% of the cases. This result indicates that the E. coli population occurring in individuals within the same household is highly diverse even when considering subpopulations characterized by a specific resistance phenotype.
Most E. coli clones occurred specifically in either humans or dogs whereas 8% of randomly obtained clones were shared between the two hosts. This value is close to the 7% reported previously by other authors [Reference Caugant, Levin and Selander24]. Although certain clones were found to occur in up to four distinct families, within-household sharing of randomly obtained clones was significantly more common than across-household sharing (P<0·001). This result corroborates similar findings by Johnson et al. [Reference Johnson4] suggesting that clone sharing is predominantly a household-specific phenomenon and that E. coli clones circulate between individuals and/or are acquired from common external sources.
A total of 19 clones were shared within six of the eight families (Table 3), and 11 of them belonged to phylotypes B2 (n=7) or D (n=4). Although transmission of clones between subjects was not proven, some interesting trends and cases were observed. A general trend was the extensive clone sharing between dogs living in the same households. Dogs B3 and B5 shared five clones throughout the study and multiple clones were also shared between the three dogs in household C and between the two dogs in household F. Clone sharing between pets living in the same household probably reflects the close physical contact between the animals but may also result from dogs being fed with table scraps. One interesting case of clone sharing occurred between a human (D3) and a dog (D5), which had lived with household D for only 2 weeks prior to the first sampling. It is likely that the dog acquired E. coli clone 147 (Table 3) upon entering the household, because this clone was isolated from the mother on the seven first sampling dates but not from the dog until the last sampling date. Because the dog was allegedly fed commercial dog food only, it can be speculated that direct or indirect transmission from the woman to the dog had occurred. Two cases of human–dog clone sharing (families A and C) involved dogs fed table scraps on a regular basis. In these cases, a common food source of E. coli can not be ruled out. The degree of direct and indirect physical contact reported between humans and dogs did not seem to influence the occurrence of clone sharing (data not shown). Hand hygiene may help in preventing transmission as suggested by the fact that the only two persons (B1 and B2) who practised hand-washing upon physical contact with their dogs did not share any clones with their dogs. However, this hypothesis was not supported by statistical evidence due to the low numbers of individuals studied.
Four of the seven clones shared between humans and dogs belonged to phylotypes B2 or D. A similar isolation frequency (two out of three isolates) of these two UTI-associated phylotypes was detected in clones shared between humans, indicating that both transmission pathways are equally important in the spread of urovirulent E. coli within households. A previous study [Reference Johnson, Stell and Delavari25] has shown that at least half of faecal E. coli from dogs possesses virulence genes associated with UTI. Dogs may also play a role in the dissemination of antimicrobial-resistant E. coli of clinical relevance [Reference Guardabassi, Schwarz and Lloyd26]. ESBL-producing E. coli have been previously isolated from both healthy and diseased dogs [Reference Carattoli27, Reference Costa28] and in this study one dog was found to shed repeatedly a CTX-M9-producing E. coli.
A limitation of this study is that only one single random isolate was typed for most samples. The inclusion of multiple isolates from each sample would certainly have increased the observed diversity, even though multiple isolates from the same faecal sample frequently represented a single clone [Reference Caugant, Levin and Selander24, Reference McLellan, Daniels and Salmore29]. We detected high diversity over time and therefore the sampling strategy did not affect our conclusion on diversity. It is more likely that such a factor may have reduced the chance of detecting clone sharing. However, this was compensated for by the use of the antimicrobial selective DPM, which allowed detection of 37% of clones shared within families. In combination with phylotyping, this approach proved to be a simple and effective method for detection of clone sharing.
In conclusion, the faecal E. coli population in most members of the eight households under study was found to be subjected to frequent temporal shifts. Although resident clones occurred in most individuals, frequent temporal shifts in the composition of the E. coli population were observed in humans and even more in dogs. Clone sharing was a common phenomenon within households and 10/18 humans studied were shown at least once to share a clone with another person or with a dog in their household. The fact that more than half of the clones shared between individuals belonged to the two phylotypes associated with UTI supports the notion that within-household transmission of urovirulent E. coli plays an important role in the pathogenesis of UTI. As previously suggested by Johnson et al. [Reference Johnson4] this epidemiological aspect has important implications for the design of preventive measures against the diffusion of ExPEC and other pathogenic or antimicrobial-resistant E. coli lineages of clinical relevance in the population.
ACKNOWLEDGEMENTS
We thank the families for their participation in this study.
NOTE
Supplementary material (details of the AFLP protocol) accompanies this paper on the Journal's website (http://journals.cambridge.org/hyg).
DECLARATION OF INTEREST
None.





