Long-chain PUFA (LCPUFA) have an essential role in fetal neurodevelopment(Reference Innis1). The LCPUFA, docosahexanoic acid (DHA, 22:6 n-3) and arachidonic acid (AA, 20:4 n-6) are of particular importance owing to their role in central nervous system development(Reference Crawford2–Reference Lauritzen, Hansen and Jorgensen6), and DHA has been shown to accumulate in the fetal brain during the third trimester(Reference Shahidi and Ambigaipalan7–Reference Makrides9). AA is essential for fetal brain growth during gestation(Reference Crawford2,Reference Crawford, Costeloe and Ghebremeskel10–Reference Tallima and El Ridi13) , whilst DHA is required for the growth and function of nervous tissue and the retina(Reference Uauy, Birch and Birch14–Reference Molloy, Doyle and Makrides17). Placental transfer is the sole source of PUFA for the developing fetus(Reference Jones, Mark and Waddell18); therefore, optimal maternal status is essential.
DHA and AA can be supplied directly to the mother from dietary intake and also synthesised endogenously from the essential fatty acids linoleic acid (LA, 18:2 n-6) and α-linolenic acid (ALA, 18:3 n-3), respectively, via a series of elongation and desaturation steps. The endogenous synthesis of LCPUFA involves the Δ 5 desaturase and Δ 6 desaturase enzymes which are encoded by the fatty acid desaturase (FADS) genes – FADS1 and FADS2, respectively(Reference Minihane19). The FADS genes are located on chromosome 11q12–q13.1(Reference Marquardt, Stöhr and White20). SNP in the FADS gene cluster are associated with LCPUFA concentrations. The minor allele homozygous genotype for FADS1 and FADS2 SNP has been associated with increased blood concentrations of the PUFA precursor molecules LA and ALA and lower concentrations of LCPUFA products, particularly AA in pregnant women(Reference de la Garza Puentes, Montes Goyanes and Chisaguano Tonato21–Reference Moltó-Puigmartí, Plat and Mensink23). Higher cord blood LA concentrations and lower concentrations of AA and DHA have been reported in children whose mothers were carriers of the minor allele for FADS1 SNP including rs3834458(Reference Steer, Hibbeln and Golding24) and rs174575 of the FADS2 gene(Reference Lattka, Koletzko and Zeilinger25). Desaturase enzyme activity has been reported in the liver of the developing fetus(Reference Chambaz, Ravel and Manier26,Reference Rodrigueza, Sarda and Nessmann27) , indicating that the endogenous synthesis of LCPUFA is active in the fetus, which may in turn be influenced by FADS genotype. The genotype of the child has also been shown to influence cord n-6 PUFA concentrations, with increased dihomo-γ-linolenic acid (DGLA)(Reference Steer, Hibbeln and Golding24,Reference Lattka, Koletzko and Zeilinger25,Reference Barman, Nilsson and Naluai28) , and decreased AA being reported for minor allele carriers of several SNP, including rs174575, rs1535(Reference Steer, Hibbeln and Golding24), rs174561, rs3834458(Reference Lattka, Koletzko and Zeilinger25), rs102275 and rs174448(Reference Barman, Nilsson and Naluai28). In a high fish-eating cohort in the Seychelles Child Development Study (SCDS), mothers who were minor allele carriers for the intergenic FADS1-FADS2 SNP rs3834458 had significantly higher serum concentrations of LA and ALA compared with major allele homozygotes, and both rs3834458 and rs174575 were associated with significantly lower concentrations of AA(Reference Yeates, Love and Engström22).
The FADS genotype, therefore, may be a determinant of circulating concentrations of PUFA in pregnant women and cord blood even in high fish-eating populations. Given that the developing fetus relies on maternal supply, it is possible that both maternal and child FADS genotype will influence cord PUFA status. The aim of the current study is to investigate the influence of maternal and child genetic variation in FADS genotype on cord blood PUFA status in the high fish-eating SCDS. It is hypothesised that carriers of minor alleles for FADS1 rs174537 and rs174561, FADS2 rs174575, and FADS1-FADS2 rs3834458 in either the mother or the child will have lower cord blood concentrations of the LCPUFA products AA and DHA.
Methods
Study design and participants
The SCDS is a longitudinal study based in the Republic of Seychelles. The SCDS has the overall aim of investigating the effects of methylmercury exposure from maternal fish consumption on child neurodevelopmental outcomes. The current study is part of Nutrition Cohort 2. Recruitment for Nutrition Cohort 2 took place between 2008 and 2011 on the island of Mahé. Pregnant women (n = 1535) were recruited at their first antenatal visit from eight health centres located on the island. The present study was conducted according to the guidelines laid down in the Declaration of Helsinki, and all procedures involving human subjects were approved by the Seychelles Ethics Board, the Research Subjects Review Board at the University of Rochester and the Regional Ethics committee at Lund University, Sweden. Written informed consent was obtained from all subjects.
Maternal characteristics
Mothers reported their age when enrolled to the study using questionnaires administered by trained nurses. BMI (BMI = weight (kg)/ height2 (m2)) was calculated using maternal weight and height measurements recorded by trained nurses when the children were approximately 20 months old. Data on BMI prenatally were not available; however, in a previous cohort (Nutrition Cohort 1), maternal postnatal BMI was found to be highly correlated (r = 0·93) with preconception BMI. Socio-economic status was determined when children were aged 20 months using Hollingshead 4-Factor Social Status Index, adapted for use in the Republic of Seychelles. Maternal fish consumption was determined using a Fish Use Questionnaire completed retrospectively by mothers to reflect fish consumption during pregnancy. The Fish Use Questionnaire allowed for total fish consumption to be estimated, as well as the consumption of oily and white fish.
Blood sampling and analyses
Maternal non-fasting blood samples, collected at 28 weeks’ gestation and cord blood samples collected at delivery, were processed following collection to obtain serum samples by centrifuging at 2500 rpm for 15 min and stored at −80 °C. Aliquots of serum were shipped to Ulster University, Coleraine, for fatty acid analysis. Total serum PUFA was assessed in cord blood samples as PUFA transferred from the mother to the developing fetus via the placenta originate from maternal triglycerides and free fatty acids, and not from phospholipids(Reference Dutta-Roy29–Reference Bonham, Duffy and Wallace31). Total PUFA analysis was completed in cord blood serum samples using an adaptation of the Folch et al. (Reference Folch, Lees and Stanley32) method. Total lipids were extracted and methylated to fatty acid methyl esters using boron trifluoride methanol (Sigma Aldrich, UK). Heptadecaenoic acid (C17:0) was used as the internal standard. Fatty acid methyl esters were quantified by GC-MS (Agilent 7890 A-5975C). Analysis was performed in split mode, with a BPX70 capillary GC column (SGE Analytical Science) (length 30 m, internal diameter 250 µm and film thickness 0·25 µm), using He as the carrier gas (constant flow at 1·0 ml/min). Samples were injected using an automatic liquid sampler (injection volume 1 µl) at a temperature of 130 °C, which was then ramped at 15 °C/min to 200 °C and then at 30 °C/min to 250 °C where it was held for 5 min. The total run time was 13 min, with all peaks for chosen PUFA resolved in this short period. MS was operated in positive ion mode using an electron ionisation source. Mass range was set to 50–500 Da and acquisition was performed by total ion chromatogram. The PUFA analysed included LA (18:2 n-6), ALA (18:3 n-3), AA (20:4 n-6), EPA (20:5 n-3) and DHA (22:6 n-3). PUFA were identified using their retention time and corresponding qualifier ions with reference to those of commercially available PUFA standards (Sigma Aldrich, UK). Quantitation was performed using the internal standard (C17:0) and PUFA quantifier ions. The sum of EPA and DHA was calculated (EPA + DHA), n-6:n-3 ratio (total n-6:total n-3), and product:precursor ratios (AA:LA, EPA:ALA and DHA:ALA) were also calculated. Undetectable values for ALA and EPA were seen in 85% and 57% of samples, respectively. These samples had values below the lower limit of detection (LLOD). For undetectable values, the LLOD/√2 was inputted(Reference Ogden33).
Genotyping
Aliquots of maternal whole blood were shipped to Lund University, Sweden, for genotyping as described previously(Reference Yeates, Love and Engström22). A total of four FADS SNP were chosen based on the impact they have previously been found to have on LCPUFA concentrations(Reference Yeates, Love and Engström22), including FADS1 rs174537, FADS1 rs174561, FADS2 rs174575 and FADS1-FADS2 intergenic rs3834458. A Qiagen DNA Blood Mini Kit (Qiagen, Germany) was used to extract DNA from maternal blood samples. Genotyping of SNP was completed using the Iplex® Gold assay on the MassARRAY platform (Sequenom™, USA) and by TaqMan allelic discrimination assay on an ABI 7900 instrument (Applied Biosciences, USA).
Children provided a saliva sample when aged 7 years old. Saliva samples were also shipped to Lund University, Sweden, for DNA extraction and genotyping. DNA extraction was completed using the E.Z.N.A Blood DNA Mini Kit (Omega Bio-Tek, USA) according to the manufacturer’s instructions. The same four SNP genotyped for maternal samples were genotyped in child saliva samples (FADS1 rs174537, FADS1 rs174561, FADS1-FADS2 intergenic rs3834458 and FADS2 rs174575). Genotyping of SNP was completed using TaqMan real-time PCR using custom assays from Thermo Fisher Scientific. To analyse the chosen SNP, ABI 7900HT Fast Real-Time PCR System (Applied Biosystems, Thermo Fisher, USA) was used according to the manufacturer’s instructions. Primers and probes were pre-designed and validated by the manufacturer (Thermo Fisher Scientific, USA). Quality control was checked by reanalysing > 5 % of random samples, with there being a resultant 100 % agreement between original and duplicate samples. Hardy–Weinberg equilibrium was also evaluated using χ 2 test.
Statistics
Statistical analysis was completed using Statistical Package for Social Sciences (SPSS), version 25 (SPSS, IBM, USA). Descriptive analysis was completed, and data were tested for normality, which indicated that PUFA status data were not normally distributed. Cord PUFA data were transformed using natural logarithmic transformation, with a constant of + 1 added. Correlation analysis was completed for maternal and cord PUFA status to investigate if maternal PUFA determines cord PUFA. Differences in cord PUFA status across genotypes were assessed using ANOVA. Log-transformed cord PUFA variables were included in all regression models. Unadjusted linear regression was used to estimate the effect of maternal and child FADS SNP on cord blood PUFA status, where child FADS and maternal FADS genotype were analysed in separate models. A further regression model was completed adding maternal FADS genotype as a covariate in order to examine more clearly the influence of child genotype on cord PUFA concentrations. Regression models were also run with maternal PUFA concentrations included as a covariate. Regression assumptions including multicollinearity (VIF), Durbin–Watson and scatterplots were examined and determined to be appropriate. Product:precursor ratios were also included in regression analyses (AA:LA, EPA:ALA and DHA:ALA). In all regression models, the major allele homozygote genotype for each SNP was used as the reference group. A P-value of P < 0·05 was considered statistically significant.
Results
A total of 1535 mothers were recruited onto Nutrition Cohort 2. Of these mothers, 1433 had PUFA concentrations measured at 28 weeks’ gestation. A total of 1088 of these mothers had a cord blood sample collected at delivery. Of those with cord blood data, 1062 had maternal FADS genotype and 916 had child FADS genotype data available. Descriptive characteristics of the mothers along with PUFA data for both mothers and children are included in Table 1. Fish consumption in this cohort of pregnant women was an average of 8·5 fish meals per week. The genotype distributions for mothers and children are shown in Online Resource 1. Both mother and child genotypes were in Hardy–Weinberg equilibrium. The minor allele frequency (MAF) of each SNP in Nutrition Cohort 2 mothers and children is shown in Online Resource 2 with comparison to other populations. The minor allele frequencies for rs174537, rs174561 and rs3834458 were lower in the Seychellois populations compared with European and global frequencies. The minor allele frequency for rs174575 was similar to that seen for African populations(34).
Table 1. Descriptive statistics for maternal and cord PUFA
(Mean values and standard deviations; median value and interquartile range)

IQR, interquartile range; Min, minimum; Max, maximum; SES, socio-economic status; LA, linoleic acid; ALA, α-linolenic acid; AA, arachidonic acid.
IQR is 25th and 75th centile.
Pearson’s correlation analysis showed maternal LA status to be significantly correlated with cord LA status (r = 0·100; P = 0·001) and maternal ALA to be significantly negatively correlated with cord AA (r = –0·091; P = 0·003). Maternal EPA + DHA, n-6:n-3 ratio and product:precursor ratios were all found to be positively correlated with cord EPA, DHA and n-3 product:precursor ratios (P < 0·05 for all) (Online Resource 3). Differences in cord PUFA concentrations according to child genotype were assessed using ANOVA and are shown in Table 2. Children who were heterozygous or homozygous carriers of the minor allele had significantly lower AA (P < 0·05 for all) concentrations in cord blood, and a significantly lower AA:LA ratio (P = 0·05 for all) in cord blood compared with those who were major allele homozygotes for all SNP. The equivalent non-parametric tests were completed on non-log-transformed data which yielded similar results.
Table 2 Cord PUFA (mg/ml) according to child FADS genotype
(Mean values and standard deviations)
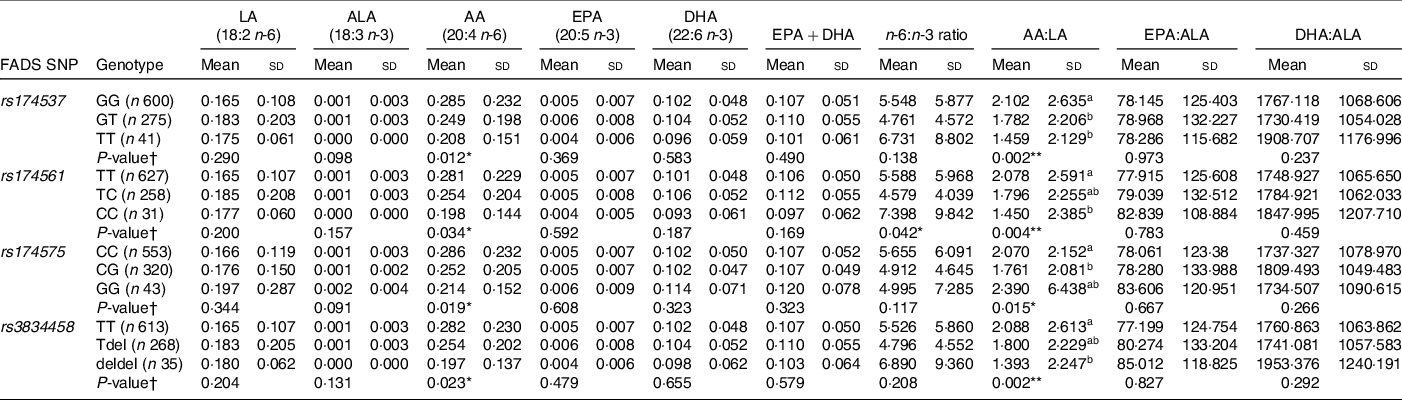
FADS, fatty acid desaturase; LA: linoleic acid; ALA: α-linolenic acid; AA: arachidonic acid.
* P < 0·05, **P < 0·01; ***P < 0·001.
† P-value for significant difference between cord PUFA concentrations according to genotype as determined using ANOVA with Tukey for post hoc comparison; all cord PUFA were logged after adding a constant to all + 1; abcmean values within a column with unlike superscript letters were significantly different (P < 0·05). AA post hoc not significant.
Regression analysis investigated associations between maternal FADS genotype and cord PUFA concentrations. Unadjusted regression models were completed, as were regression models including maternal PUFA concentrations as a covariate. The results of these analyses were the same, albeit the level of significance may have changed slightly in some instances. As the same results were significant, the results from unadjusted models have been reported (Table 3). In mothers, minor allele homozygotes for the FADS1 (rs174537 and rs174561) and FADS1-FADS2 (r3834458) SNP were associated with lower cord DHA (P < 0·05 for all) and lower EPA + DHA (P < 0·05 for all) when compared with the reference genotype. The heterozygous genotype was associated with increased concentrations of precursor molecules, specifically LA, when compared with major allele homozygotes for rs174561 (P = 0·013) and rs3834458 (P = 0·013). The homozygous minor allele for FADS2 rs174575 was also associated with increased LA when compared with the reference genotype (P = 0·002).
Table 3 Associations between cord PUFA concentrations (mg/ml) and maternal FADS genotype
(Coefficient values and 95 % confidence intervals)

FADS, fatty acid desaturase; LA: linoleic acid; ALA: α-linolenic acid; AA: arachidonic acid.
All cord PUFA were logged after adding a constant to all + 1; homozygous major allele genotype was reference to which heterozygous and minor allele homozygous were compared; β-values displayed are for the heterozygous genotype or minor allele homozygous compared with major allele homozygous genotype.
* P < 0·05, **P < 0·01; ***P < 0·001.
Associations between child FADS genotype and cord PUFA concentrations are shown in Table 4. The homozygous minor allele genotype for rs3834458 was associated with significantly lower AA concentrations when compared with the major allele homozygotes (P = 0·037). The data suggest a trend for lower AA concentrations for minor allele homozygotes for rs174537 (P = 0·053), rs174561 (P = 0·053) and rs174575 (P = 0·085), albeit this was not significant. A lower cord AA:LA ratio was also observed for children who were heterozygotes for rs174537, rs174561 and rs174575 (P < 0·05 for all), and also in those who were minor allele homozygotes for rs174537, rs174561 and rs3834458 (P < 0·05 for all).
Table 4 Associations between cord PUFA concentrations (mg/ml) and child FADS genotype
(Coefficient values and 95 % confidence intervals)

FADS, fatty acid desaturase; LA: linoleic acid; ALA: α-linolenic acid; AA: arachidonic acid.
All cord PUFA were logged after adding a constant to all + 1; homozygous major allele genotype was reference to which heterozygous and minor allele homozygous were compared; β-values displayed are for the heterozygous genotype or minor allele homozygous compared with major allele homozygous genotype.
* P < 0·05, **P < 0·01; P < 0·001.
When the influence of maternal FADS genotype was taken into account (Table 5), significant associations were observed between cord ALA concentrations and child genotype, where being homozygous for the minor allele for FADS1 rs174537, rs174561 and FADS1-FADS2 rs3834458 was associated with lower concentrations of ALA. This finding was also observed in children who were heterozygous for FADS2 rs174575 (P = 0·025). There were significant associations among children being heterozygous for FADS1 rs174561 and higher cord concentrations of DHA, EPA + DHA and lower n-6:n-3 ratio (P = 0·021; P = 0·027; P = 0·004, respectively), whereas there were no associations among children being homozygotes for these outcomes. The n-6:n-3 ratio was also significantly lower in children who were heterozygous for FADS1 rs174537 and FADS1-FADS2 rs3834458 (P = 0·019; P = 0·028, respectively).
Table 5 Associations between cord PUFA concentrations (mg/ml) and child FADS genotype whilst adjusting for maternal genotype
(Coefficient values and 95 % confidence intervals)
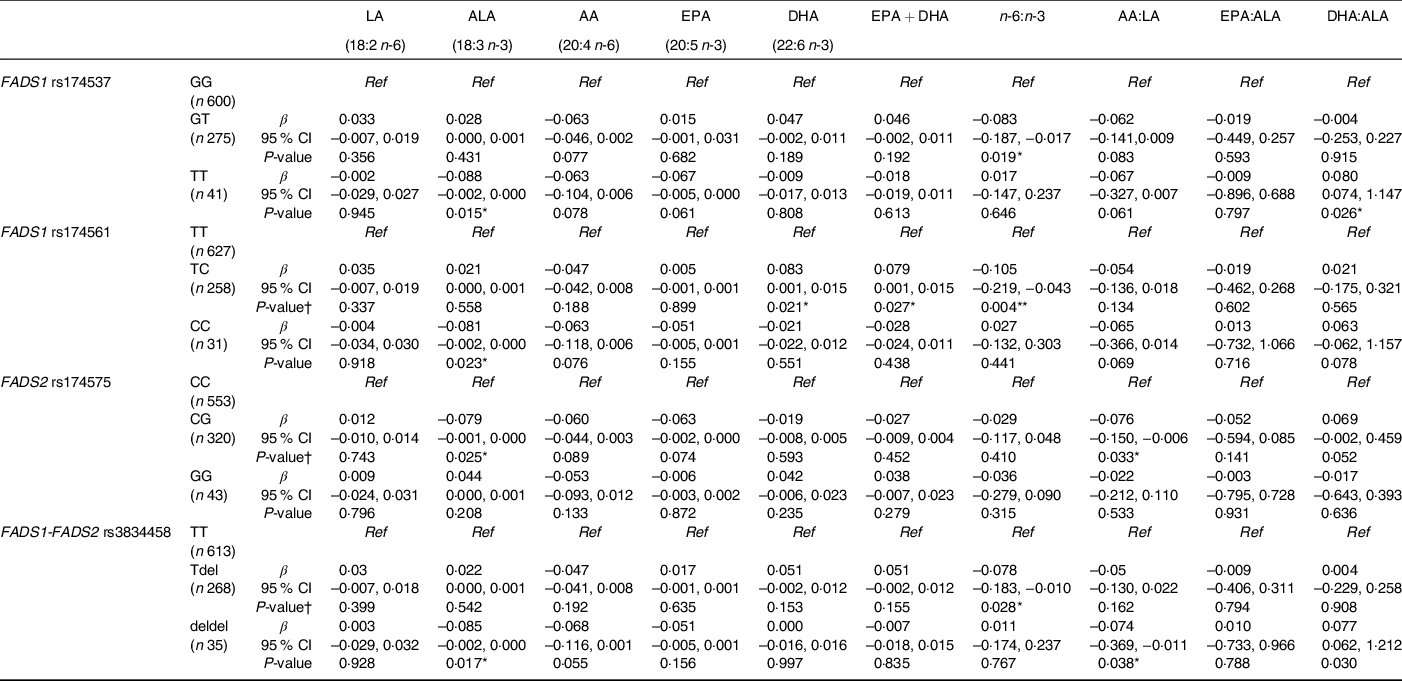
FADS, fatty acid desaturase; LA: linoleic acid; ALA: α-linolenic acid; AA: arachidonic acid.
All cord PUFA were logged after adding a constant to all + 1; homozygous major allele genotype was reference to which heterozygous and minor allele homozygous were compared; β-values displayed are for the heterozygous genotype or minor allele homozygous compared with major allele homozygous genotype; maternal FADS genotype was adjusted for in each regression model.
* P < 0·05, **P < 0·01; P < 0·001.
Discussion
The present study indicates that both maternal and child FADS genotype are associated with cord PUFA status in a high fish-eating cohort. Cord DHA concentrations were found to be influenced by maternal genotype, but not by child genotype, which was primarily associated with lower AA or ALA concentrations. FADS1 and FADS2 appear to behave differently, specifically with FADS1 rs174537 and rs174561 found to influence DHA and AA status, whereas this was not seen for the FADS2 rs174575.
The presence of the minor allele in the maternal genotype was associated with increased LA for FADS1 rs174561, and lower cord blood DHA and EPA + DHA concentrations for FADS1 rs174537, rs174575 and rs3834458. Previous research in a cohort of mother–child pairs in England examined the influence of maternal FADS on cord PUFA concentrations and also reported lower cord blood DHA concentrations in children born to mothers who were carriers of the minor allele for various FADS1 and FADS2 SNP(Reference Steer, Hibbeln and Golding24,Reference Lattka, Koletzko and Zeilinger25) . The total LCPUFA concentrations in cord blood are generally believed to be higher than maternal LCPUFA status in what is known as ‘biomagnification’(Reference Crawford, Hassam and Williams35). The placenta has the ability to select and transfer AA and DHA from the maternal supply to the fetus(Reference Crawford, Costeloe and Ghebremeskel10,Reference Crawford36) , while also retaining the precursor molecules LA and ALA(Reference Crawford, Costeloe and Ghebremeskel10). In the current analysis, cord DHA was shown to be lower in children whose mothers were minor allele carriers for FADS1 rs174537, rs174561 and FADS1-FADS2 rs3834458, suggesting that although there is a preferential transfer and biomagnification of DHA, maternal FADS genotype may influence the quantity of DHA present in fetal cord blood. The order of transfer of LCPUFA is dependent on maternal status. When maternal AA status is low, it is suggested that the placenta will retain AA and transfer PUFA to the fetus in the order DHA, ALA, LA and lastly AA(Reference Haggarty37). If, however, maternal status of AA is high, the order of selectivity and thus transfer to the fetus is DHA, AA, ALA followed by LA(Reference Haggarty37). The maternal PUFA status of this cohort has previously been shown to be influenced by maternal FADS genotype, in that maternal AA concentrations were decreased with minor allele homozygosity for rs3834458(Reference Yeates, Love and Engström22). In the current study, we have confirmed that children who are homozygous for rs3834458 also have lower AA concentrations. We have added to these findings to show that the genotype of the mother, but not the child, influences cord DHA concentrations, which may suggest that the mother’s genotype is influential in determining the order of placental transfer of PUFA to the fetus.
The endogenous synthesis of LCPUFA is active in the fetus despite maternal transfer of LCPUFA being important for fetal status. In the developing fetus, the endogenous synthesis of AA is known to be greater than the synthesis of DHA(Reference Haggarty37,Reference Lapillonne and Moltu38) . Variation in child FADS genotype influences the endogenous synthesis of LCPUFA, with lower cord blood concentrations of AA in those with the minor allele(Reference Steer, Hibbeln and Golding24,Reference Lattka, Koletzko and Zeilinger25,Reference Barman, Nilsson and Naluai28) . In the current analysis for child genotype, rs3834458 were associated with lower cord blood AA concentrations; this has been observed previously in the limited research relating to the influence of FADS on cord PUFA status(Reference Steer, Hibbeln and Golding24,Reference Lattka, Koletzko and Zeilinger25,Reference Barman, Nilsson and Naluai28) . This finding suggests that the presence of the minor allele may impair the developing fetus’ ability to endogenously synthesise LCPUFA, leading to lower cord AA concentrations.
Child genotype was shown to be associated with product:precursor ratios, with a decreased cord blood AA:LA ratio in children who were heterozygous for rs174537, rs174561 and rs3834458. Minor allele homozygous children for rs174537, rs174561 and rs3834458 also had a lower AA:LA ratio compared to children with the reference genotype, indicating decreased activity of the desaturase enzymes and thus leading to lower concentrations of AA in cord blood compared to LA. The presence of the minor allele is known to be associated with increased LCPUFA precursor and lower product concentrations. Given the inefficiency of the LCPUFA endogenous synthesis pathway, dietary intake of preformed LCPUFA is preferred. It should be noted that when adjusting for maternal genotype, some associations were found for child genotype and DHA, but only among heterozygotes and not in the expected direction. Similar unexpected associations were found between the child homozygous genotype and lower ALA. As this is the opposite to what is expected with lower FADS1 and FADS2 enzyme activity, these findings should be interpreted carefully. The present study was completed in a high fish-eating cohort, with a diet rich in preformed LCPUFA. Participants in the Seychelles are consuming on average 8·5 fish meals per week, which is much higher than the mean weekly fish consumption of other populations such as the UK(Reference Bates, Cox and Nicholson39) and USA(Reference Jahns, Raatz and Johnson40). The maternal concentrations of EPA and DHA reported for Seychellois pregnant women are reflective of this high fish consumption and are notably higher than other populations with reported fish intake which is much lower such as in the UK(Reference Moon, Harvey and Robinson41) and the USA(Reference Vidakovic, Gishti and Voortman42). Despite the high intake of preformed LCPUFA, associations between maternal and child FADS genotype and cord PUFA were observed in the current analysis, suggesting that the pathway may be more important in optimal fetal PUFA status than previously believed.
To our knowledge, this is the first study to investigate the influence of both maternal and child FADS genotype on cord PUFA status in a high fish-eating cohort. The present study has some limitations. There are other potential mechanisms aside from desaturation of PUFA by which FADS variation has an influence on PUFA status owing to changes in FADS gene expression. These mechanisms include altered promoter region of the FADS genes(Reference Brenna, Kothapalli and Park43,Reference Rahbar, Ainsworth and Howard44) and low expression of protein(Reference Ralston, Matravadia and Gaudio45), and these other potential mechanisms should be considered in future studies. We measured total serum PUFA concentrations to account for triacylglycerols which are known to be the major lipid fraction transferred from the mother to the fetus(Reference Herrera30). However, it is acknowledged that measuring other lipid fractions, such as plasma fatty acids, may yield different results. Future analysis could also consider using the omega-3 index as a measure of PUFA status in order to be more comparable to other studies. Further investigation of other genotypes should also be considered, such as variation in ELOVL genes, which are involved in the elongation steps of the endogenous synthesis of LCPUFA(Reference Barman, Nilsson and Naluai28).
Conclusion
In conclusion, the current study indicates that in a high fish-eating cohort, genetic variation in both maternal and child FADS genes influences cord PUFA concentrations, but for different PUFA. Maternal genotype was associated with lower cord DHA and EPA + DHA concentrations, whereas child genotype was associated with lower cord ALA and AA concentrations. The cohort included here have high fish consumption; however, despite the high dietary intake of preformed LCPUFA, both the maternal and child FADS genotype were shown to be associated with cord PUFA status. Given the importance of PUFA for child neurodevelopment, it is necessary to understand factors which impact PUFA concentrations and to use this knowledge to improve PUFA status of both the mother and child. Further research is needed to determine whether increased dietary intake can compensate for lower PUFA status as a result of FADS genotype.
Acknowledgements
We gratefully acknowledge the participation of all women and children who took part in the study and the nursing staff from the Child Development Centre, Seychelles, for their assistance with data collection.
This research was supported by the Department for Employment and Learning (DEL), Northern Ireland; the US National Institutes of Health (NIH) (grant numbers R01-ES010219 and P30-ES01247); the Swedish Research Council FORMAS and the Karolinska Institutet, Sweden. The study funders had no role in the design, analysis or writing of this article.
G. J. M., P. W. D., E. vW., C. F. S. and J. J. S. conceived and designed the SCDS and conducted the research. M. C. C., A. J. Y., M. S. M., E. M. M., K. W., K. B. and D. P. conducted the research. K. B., D. P. and K. W. conducted genotyping analysis. M. C. C., T. S. and M. W. conducted cord blood PUFA analysis. B. W. H. and D. F. C. provided expert advice on GC-MS. M. C. C. and M. S. M. conducted the statistical analysis. M. C. C. drafted the manuscript, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
No authors have any conflicts of interest to declare.
Supplementary material
For supplementary material/s referred to in this article, please visit https://doi.org/10.1017/S0007114521000441








