Introduction
Considerable effort has been made over the last 30 years to identify biological and cognitive markers of schizophrenia. A large number of studies have reported significant differences in chronic schizophrenia (ChSz) patients relative to healthy controls (HCs) across a range of neurobiological and neurocognitive measures. These include structural magnetic resonance imaging (sMRI), functional MRI (fMRI) and diffusion tensor MRI (DTI) (Ellison-Wright et al. Reference Ellison-Wright, Glahn, Laird, Thelen and Bullmore2008; Ellison-Wright & Bullmore, Reference Ellison-Wright and Bullmore2009; Minzenberg et al. Reference Minzenberg, Laird, Thelen, Carter and Glahn2009; Pettersson-Yeo et al. Reference Pettersson-Yeo, Allen, Benetti, McGuire and Mechelli2011), genotype (Ripke et al. Reference Ripke, Sanders, Kendler, Levinson, Sklar, Holmans, Lin, Duan, Ophoff, Andreassen, Scolnick, Cichon, St Clair, Corvin, Gurling, Werge, Rujescu, Blackwood, Pato, Malhotra, Purcell, Dudbridge, Neale, Rossin, Visscher, Posthuma, Ruderfer, Fanous, Stefansson, Steinberg, Mowry, Golimbet, De Hert, Jönsson, Bitter, Pietiläinen, Collier, Tosato, Agartz, Albus, Alexander, Amdur, Amin, Bass, Bergen, Black, Børglum, Brown, Bruggeman, Buccola, Byerley, Cahn, Cantor, Carr, Catts, Choudhury, Cloninger, Cormican, Craddock, Danoy, Datta, de Haan, Demontis, Dikeos, Djurovic, Donnelly, Donohoe, Duong, Dwyer, Fink-Jensen, Freedman, Freimer, Friedl, Georgieva, Giegling, Gill, Glenthøj, Godard, Hamshere, Hansen, Hansen, Hartmann, Henskens, Hougaard, Hultman, Ingason, Jablensky, Jakobsen, Jay, Jürgens, Kahn, Keller, Kenis, Kenny, Kim, Kirov, Konnerth, Konte, Krabbendam, Krasucki, Lasseter, Laurent, Lawrence, Lencz, Lerer, Liang, Lichtenstein, Lieberman, Linszen, Lönnqvist, Loughland, MacLean, Maher, Maier, Mallet, Malloy, Mattheisen, Mattingsdal, McGhee, McGrath, McIntosh, McLean, McQuillin, Melle, Michie, Milanova, Morris, Mors, Mortensen, Moskvina, Muglia, Myin-Germeys, Nertney, Nestadt, Nielsen, Nikolov, Nordentoft, Norton, Nöthen, O'Dushlaine, Olincy, Olsen, O'Neill, Orntoft, Owen, Pantelis, Papadimitriou, Pato, Peltonen, Petursson, Pickard, Pimm, Pulver, Puri, Quested, Quinn, Rasmussen, Réthelyi, Ribble, Rietschel, Riley, Ruggeri, Schall, Schulze, Schwab, Scott, Shi, Sigurdsson, Silverman, Spencer, Stefansson, Strange, Strengman, Stroup, Suvisaari, Terenius, Thirumalai, Thygesen, Timm, Toncheva, van den Oord, van Os, van Winkel, Veldink, Walsh, Wang, Wiersma, Wildenauer, Williams, Williams, Wormley, Zammit, Sullivan, O'Donovan, Daly and Gejman2011; Steinberg et al. Reference Steinberg, de Jong, Andreassen, Werge, Børglum, Mors, Mortensen, Gustafsson, Costas, Pietiläinen, Demontis, Papiol, Huttenlocher, Mattheisen, Breuer, Vassos, Giegling, Fraser, Walker, Tuulio-Henriksson, Suvisaari, Lönnqvist, Paunio, Agartz, Melle, Djurovic, Strengman, Jürgens, Glenthøj, Terenius, Hougaard, Ørntoft, Wiuf, Didriksen, Hollegaard, Nordentoft, van Winkel, Kenis, Abramova, Kaleda, Arrojo, Sanjuán, Arango, Sperling, Rossner, Ribolsi, Magni, Siracusano, Christiansen, Kiemeney, Veldink, van den Berg, Ingason, Muglia, Murray, Nöthen, Sigurdsson, Petursson, Thorsteinsdottir, Kong, Rubino, De Hert, Réthelyi, Bitter, Jönsson, Golimbet, Carracedo, Ehrenreich, Craddock, Owen, O'Donovan, Ruggeri, Tosato, Peltonen, Ophoff, Collier, St Clair, Rietschel, Cichon, Stefansson, Rujescu and Stefansson2011) and neuropsychological profile (Tyson et al. Reference Tyson, Laws, Roberts and Mortimer2004; Minzenberg et al. Reference Minzenberg, Laird, Thelen, Carter and Glahn2009). More recently, efforts to facilitate earlier and more effective treatment intervention have resulted in studies focusing on those in the earliest stages of the illness, namely, individuals with first-episode psychosis (FEP) and those deemed to be at ultra-high risk (UHR). In these groups, similar neuroanatomical, neurofunctional and cognitive alterations (Bilder et al. Reference Bilder, Goldman, Robinson, Reiter, Bell, Bates, Pappadopulos, Willson, Alvir, Woerner, Geisler, Kane and Lieberman2000; Keefe et al. Reference Keefe, Perkins, Gu, Zipursky, Christensen and Lieberman2006; Wood et al. Reference Wood, Brewer, Koutsouradis, Phillips, Francey, Proffitt, Yung, Jackson, McGorry and Pantelis2007; Walterfang et al. Reference Walterfang, Yung, Wood, Reutens, Phillips, Wood, Chen, Velakoulis, McGorry and Pantelis2008; Benetti et al. Reference Benetti, Mechelli, Picchioni, Broome, Williams and McGuire2009; Crossley et al. Reference Crossley, Mechelli, Fusar-Poli, Broome, Matthiasson, Johns, Bramon, Valmaggia, Williams and McGuire2009; Allen et al. Reference Allen, Stephan, Mechelli, Day, Ward, Dalton, Williams and McGuire2010, Reference Allen, Chaddock, Howes, Egerton, Seal, Fusar-Poli, Valli, Day and McGuire2011; Koutsouleris et al. Reference Koutsouleris, Patschurek-Kliche, Scheuerecker, Decker, Bottlender, Schmitt, Rujescu, Giegling, Gaser, Reiser, Möller and Meisenzahl2010; Seidman et al. Reference Seidman, Giuliano, Meyer, Addington, Cadenhead, Cannon, McGlashan, Perkins, Tsuang, Walker, Woods, Bearden, Christensen, Hawkins, Heaton, Keefe, Heinssen and Cornblatt2010; Mechelli et al. Reference Mechelli, Riecher-Rössler, Meisenzahl, Tognin, Wood, Borgwardt, Koutsouleris, Yung, Stone, Phillips, McGorry, Valli, Velakoulis, Woolley, Pantelis and McGuire2011; Fusar-Poli et al. Reference Fusar-Poli, Smieskova, Serafini, Politi and Borgwardt2012) that may be mediated genetically (Fusar-Poli et al. Reference Fusar-Poli, Smieskova, Serafini, Politi and Borgwardt2012) have also been reported, though such alterations are usually less severe than those seen in ChSz groups (Egerton et al. Reference Egerton, Borgwardt, Tognin, Howes, McGuire and Allen2011). The majority of studies, however, have largely employed univariate analyses that allow inference at the group level only. Therefore, to promote the clinical translation of such work, efforts have progressively turned toward alternative analytical approaches that allow inference at the level of the individual.
One such technique is the supervised learning method support vector machine (SVM). A type of multivariate pattern recognition algorithm, SVM has become increasingly used in studies of psychiatric and neurological disorder (Orrù et al. Reference Orrù, Pettersson-Yeo, Marquand, Sartori and Mechelli2012), the rationale for which is twofold: first, SVM allows inference at the single-subject, rather than group, level (Lao et al. Reference Lao, Shen, Xue, Karacali and Resnick2004; Norman et al. Reference Norman, Polyn, Detre and Haxby2006); second, as a multivariate analysis, SVM is able to account for the inter-relationship between different within-modality measures for each subject by considering them simultaneously. Specifically, SVM involves the development of a generalized decision function [represented by an optimal separating hyperplane (OSH)] using a known ‘training’ dataset (e.g. voxel intensities), able to discriminate between examples (i.e. subjects) belonging to two predefined classes (e.g. diagnostic categories). This function is then applied to new, as yet unseen ‘test’ data, and its accuracy assessed in terms of the proportion of examples correctly classified providing an estimate of how well the classifier can be expected to generalize to future individual cases (Pereira et al. Reference Pereira, Mitchell and Botvinick2009).
At present, only a handful of studies have used SVM to investigate psychosis, with those that have predominantly employing data from only a single modality. Studies using sMRI, for example, report that ChSz patients can be significantly discriminated from HCs with accuracies of 81.1% (Davatzikos et al. Reference Davatzikos, Shen, Gur, Wu, Liu, Fan, Hughett, Turetsky and Gur2005) and 86.1% (Sun et al. Reference Sun, van Erp, Thompson, Bearden, Daley, Kushan, Hardt, Nuechterlein, Toga and Cannon2009), and UHR subjects from HCs with an accuracy of 82% (Koutsouleris et al. Reference Koutsouleris, Meisenzahl, Davatzikos, Bottlender, Frodl, Scheuerecker, Schmitt, Zetzsche, Decker, Reiser, Möller and Gaser2009a). DTI has also been used by one study which reported that ChSz patients can be discriminated from HCs with 90.62% accuracy based on white matter (WM) integrity (Ingalhalikar et al. Reference Ingalhalikar, Kanterakis, Gur, Roberts and Verma2010). In addition two studies using fMRI data reported the ability to discriminate ChSz patients from HCs with accuracies of 92.4% (Costafreda et al. Reference Costafreda, Fu, Picchioni, Toulopoulou, McDonald, Kravariti, Walshe, Prata, Murray and McGuire2011) and 81.6% (Yang et al. Reference Yang, Liu, Sui, Pearlson and Calhoun2010), respectively. By contrast, only one study has applied SVM to genotype data in the context of psychosis, and reported that genetic information could accurately discriminate ChSz subjects from HCs with 73.9% accuracy (Yang et al. Reference Yang, Liu, Sui, Pearlson and Calhoun2010). Lastly, cognitive profile has also been employed by one recent study which reported that UHR subjects were discriminable from HCs with 94.2% accuracy (Koutsouleris et al. Reference Koutsouleris, Davatzikos, Bottlender, Patschurek-Kliche, Scheuerecker, Decker, Gaser, Möller and Meisenzahl2011). Taken together, these studies support the notion that the individual use of different modalities, each in conjunction with SVM, may allow discrimination between those at different stages of the psychosis time-course at the single-subject level. However, as no investigation has yet gathered data from such a wide range of modalities during the same study, the relative accuracies of genetic, DTI, sMRI, fMRI and neuropsychological data within the same population(s) are unknown. Furthermore, since the majority of work so far has applied predominantly to those with ChSz, it is less clear whether these same metrics can be reliably utilized to draw inference at the individual level for FEP and UHR subjects, differentiating them either from HCs, or, for the first time at cross-sectional level, from each other.
In the current study, therefore, we aimed to investigate the discriminative potential of genetic, sMRI, fMRI, DTI and/or cognitive data in the classification of FEP, UHR and HC subjects at the individual level. Based on the evidence currently available, our hypotheses were threefold. (1) FEP subjects would be discriminable from HCs across the largest range of modalities, since at the group level greater magnitudes of difference have been reported in FEP over UHR subjects relative to HCs (Bilder et al. Reference Bilder, Goldman, Robinson, Reiter, Bell, Bates, Pappadopulos, Willson, Alvir, Woerner, Geisler, Kane and Lieberman2000; Eastvold et al. Reference Eastvold, Heaton and Cadenhead2007; Fusar-Poli et al. Reference Fusar-Poli, Howes, Valli, Allen, Broome, Grasby and McGuire2009, Reference Fusar-Poli, Smieskova, Serafini, Politi and Borgwardt2012; de Mello Ayres et al. Reference de Mello Ayres, Scazufca, Menezes, Nakano, Regina, Schaufelberger, Murray, McGuire, Rushe and Busatto2010; Smieskova et al. Reference Smieskova, Fusar-Poli, Allen, Bendfeldt, Stieglitz, Drewe, Radue, McGuire, Riecher-Rössler and Borgwardt2010). (2) In comparison with FEP subjects, UHR subjects would be differentiable from HCs by fewer modalities, with sMRI, DTI, fMRI and cognitive data being the most sensitive measures, in correspondence with previous group-level results (Pflueger et al. Reference Pflueger, Gschwandtner, Stieglitz and Riecher-Rössler2007; Benetti et al. Reference Benetti, Mechelli, Picchioni, Broome, Williams and McGuire2009; Crossley et al. Reference Crossley, Mechelli, Fusar-Poli, Broome, Matthiasson, Johns, Bramon, Valmaggia, Williams and McGuire2009; Koutsouleris et al. Reference Koutsouleris, Schmitt, Gaser, Bottlender, Scheuerecker, McGuire, Burgermeister, Born, Reiser, Möller and Meisenzahl2009b). (3) We would be able to directly discriminate FEP from UHR subjects, with sMRI and cognitive data being the most able to differentiate the two groups (Riecher-Rössler et al. Reference Riecher-Rössler, Pflueger, Aston, Borgwardt, Brewer, Gschwandtner and Stieglitz2009; Mechelli et al. Reference Mechelli, Riecher-Rössler, Meisenzahl, Tognin, Wood, Borgwardt, Koutsouleris, Yung, Stone, Phillips, McGorry, Valli, Velakoulis, Woolley, Pantelis and McGuire2011).
Method
A brief description of the materials and methods is provided in this section. For further methodological details, please see Supplement 1.
Subjects
FEP
A total of 19 subjects were recruited through the South London and Maudsley National Health Service Trust (http://www.slam.nhs.uk). All had experienced a FEP within the past 24 months that met Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV) criteria for a schizophreniform psychosis.
At-risk mental state
A total of 19 subjects were recruited from Outreach and Support in Southeast London (OASIS), a clinical service for young people at high risk of developing psychosis (Broome et al. Reference Broome, Woolley, Johns, Valmaggia, Tabraham, Gafoor, Bramon and McGuire2005). Their clinical status was defined according to the Personal Assessment and Crisis Evaluation (PACE) criteria (Yung et al. Reference Yung, Phillips, McGorry, McFarlane, Francey, Harrigan, Patton and Jackson1998) and their diagnosis confirmed using the Comprehensive Assessment of At-Risk Mental States (CAARMS; Yung et al. Reference Yung, Yuen, McGorry, Phillips, Kelly, Dell'Olio, Francey, Cosgrave, Killackey, Stanford, Godfrey and Buckby2005). In brief, individuals are classed as being UHR based on the presence of (1) attenuated psychotic symptoms, (2) brief limited intermittent psychotic symptoms, or (3) trait and state risk factors (e.g. individual has a schizotypal personality disorder or a first-degree relative with a DSM-IV psychiatric disorder combined with a significant decline in cognitive and social functioning over the past year).
For FEP and UHR subjects, psychopathology was measured on the day of scanning using the Positive and Negative Syndrome Scale (PANSS; Kay et al. Reference Kay, Opler and Fiszbein1987).
HCs
A total of 23 subjects were recruited from the local area through advertising. No subjects met criteria for a DSM-IV psychiatric disorder, fulfilled the PACE criteria for prodromal symptoms or had a first-degree family history of psychiatric disorder.
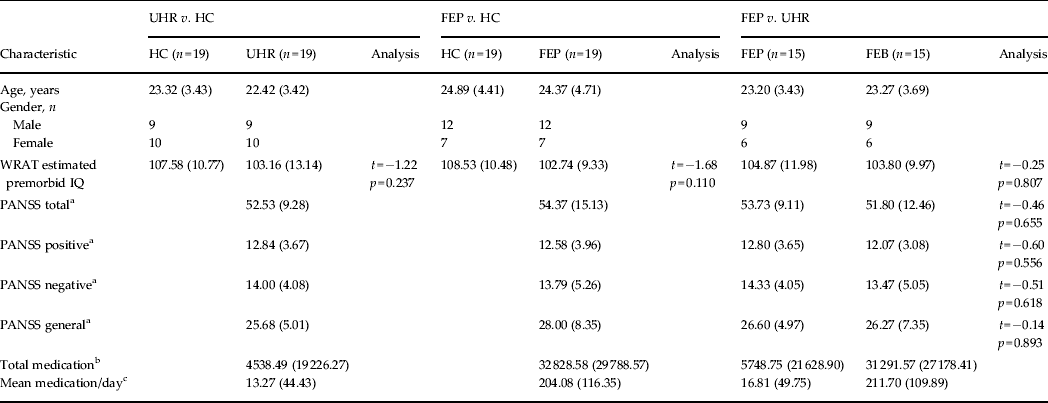
All subjects included in the study were aged 18–35 years and spoke English as their first language. Exclusion criteria included a history of neurological disorder, DSM-IV criteria for substance misuse disorder, or prior head trauma resulting in loss of consciousness and/or hospitalization (Table 1).
Table 1. Demographic data for each SVM diagnostic comparison

Data are given as mean (s.d.).
SVM, Support vector machine; UHR, ultra-high risk; HC, healthy control; FEP, first-episode psychosis; WRAT, Wide Range Achievement Test; PANSS, Positive and Negative Syndrome Scale; s.d., standard deviation.
a Symptom profile recorded at the time of the scan.
b Total medication refers to the average absolute amount of medication taken by that group in standardized mg units of chlorpromazine ± 1 s.d.
c Mean medication/day is the average medication dosage taken by each subject during their period of treatment in standardized mg units of chlorpromazine ± 1 s.d.
Data acquisition
MRI
All neuroimaging was conducted using a 3-T MRI scanner (Sigma LX-GE, USA) at the Maudsley Hospital, London. For sMRI-derived grey matter (GM) images, T1-weighted scans were obtained with a volumetric three-dimensional Spoiled Gradient Recall sequence [repetition time (TR) = 7.044 ms, echo time (TE) = 2.82 ms, flip angle = 20°, slice thickness = 1.1 mm, in-plane resolution = 1.09 × 1.09 mm, field of view (FOV) = 21 cm2, matrix = 256 × 256] producing 196 coronal slices. For DTI-derived fractional anisotropy (FA) maps, volumes were acquired using a multi-slice peripherally-gated doubly refocused spin-echo echo planar imaging (EPI) sequence, optimized for precise measurement of the diffusion tensor in parenchyma, from 60 contiguous near-axial slice locations with a TE = 104.5 ms, flip angle = 90°, slice thickness = 2.4 mm, FOV = 30.7 cm2 and matrix = 128 × 128. The maximum diffusion was 1300 s/mm2 and four images were acquired at slice locations with no diffusion gradients, alongside 32 diffusion-weighted images in which gradient directions were uniformly distributed in space. Functional images were acquired using a TR = 2000 ms, TE = 30 ms, flip angle = 70°, slice thickness = 3 mm, FOV = 24 cm2 and matrix = 64 × 64 producing 38 axial slices in parallel to the AC-PC (anterior commissure–posterior commissure) line. During the acquisition of functional images, subjects performed the Hayling sentence completion task (HSCT) using an experimental protocol described elsewhere (Allen et al. Reference Allen, Mechelli, Stephan, Day, Dalton, Williams and McGuire2008). In brief, subjects were visually presented for 4 s with a five-, six- or seven-word sentence-stem with the last word omitted. Presentation of a question mark then required them to overtly generate a word either congruent (initiation condition) or incongruent (suppression condition) with the preceding sentence. The task was arranged into eight blocks of five sentence-stems, with each block separated by a baseline condition whereby the subject was shown a visual fixation-cross for 4 s, followed by the word ‘REST’ for 4 s which they had to read overtly. Overall, one initiation session and one suppression session were run separately, generating 600 image volumes in total.
Molecular genetics
Saliva samples were obtained from each subject using the Oragene® DNA collection kit (DNA Genotek Inc., Canada), preceded by 30 min of nil by mouth.
Neuropsychology
Designed to quantify different components of verbal learning, retention and retrieval (Delis et al. Reference Delis, Kramer, Kaplan and Ober1987) the California Verbal Learning Test, Second Edition (CVLT-II) is a neuropsychological test that comes provided with associated demographically corrected norms (Delis et al. Reference Delis, Kramer, Kaplan and Ober2000). This test was chosen since it had revealed robust deficits in ChSz patients, FEP and UHR subjects relative to matched controls in previous studies (Cirillo & Seidman, Reference Cirillo and Seidman2003; Rund et al. Reference Rund, Melle, Friis, Larsen, Midbøe, Opjordsmoen, Simonsen, Vaglum and McGlashan2004; Niendam et al. Reference Niendam, Bearden, Johnson, McKinley, Loewy, O'Brien, Nuechterlein, Green and Cannon2006). Prior to scanning, the CVLT-II was administered to each subject by a trained researcher and their answers recorded.
Data analysis
sMRI
Structural images were pre-processed using the Diffeomorphic Anatomical Registration using Exponentiated Lie algebra (DARTEL) toolbox (Ashburner, Reference Ashburner2007) in SPM8 software (http://www.fil.ion.ucl.ac.uk/spm) running under Matlab7.1 (Math Works, USA). This procedure involves the creation of a study-specific template and the segmentation of each individual image using said template, with the aim of maximizing accuracy and sensitivity.
DTI
The diffusion data were pre-processed using the ExploreDTI (Leemans et al. Reference Leemans, Jeurissen, Sijbers and Jones2009) software package, including the RESTORE (robust estimation of tensors by outlier rejection) algorithm (Chang et al. Reference Chang, Jones and Pierpaoli2005), in order to generate FA maps corrected for eddy current distortion, head motion, b-matrix reorientation and rejection of data outliers. These images were then used to create FA ‘skeletons’ depicting each subject's unique WM network and associated FA value defined integrity for each voxel, using tract-based spatial statistics (Smith et al. Reference Smith, Jenkinson, Johansen-Berg, Rueckert, Nichols, Mackay, Watkins, Ciccarelli, Cader, Matthews and Behrens2006) software.
fMRI
Functional images were pre-processed using SPM8 software (http://www.fil.ion.ucl.ac.uk/spm) running under Matlab7.1 (Math Works, USA). Following the standard SPM8 functional imaging pipeline for pre-processing and analysis, using the parameter estimates obtained for all brain voxels from the task's six experimental conditions: (1) initiation (In); (2) suppression (Su); (3) repetition of ‘REST’ during initiation (RI); (4) repetition of ‘REST’ during suppression (RS); (5) cross-fixation during initiation (CFI); and (6) cross-fixation during suppression (CFS), five contrasts of interest were computed, namely, Su > In, Su > RS, In > RI, Su > CFS and In > CFI.
Genotyping
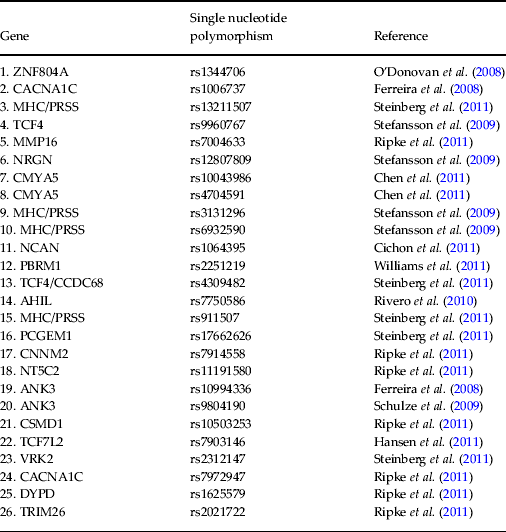
DNA was extracted from saliva samples and genotyped for a pre-selected list of 26 psychosis-associated single nucleotide polymorphisms (SNPs) (Ferreira et al. Reference Ferreira, O'Donovan, Meng, Jones, Ruderfer, Jones, Fan, Kirov, Perlis, Green, Smoller, Grozeva, Stone, Nikolov, Chambert, Hamshere, Nimgaonkar, Moskvina, Thase, Caesar, Sachs, Franklin, Gordon-Smith, Ardlie, Gabriel, Fraser, Blumenstiel, Defelice, Breen, Gill, Morris, Elkin, Muir, McGhee, Williamson, MacIntyre, MacLean, St Clair, Robinson, Van Beck, Pereira, Kandaswamy, McQuillin, Collier, Bass, Young, Lawrence, Ferrier, Anjorin, Farmer, Curtis, Scolnick, McGuffin, Daly, Corvin, Holmans, Blackwood, Gurling, Owen, Purcell, Sklar and Craddock2008; O'Donovan et al. Reference O'Donovan, Craddock, Norton, Williams, Peirce, Moskvina, Nikolov, Hamshere, Carroll, Georgieva, Dwyer, Holmans, Marchini, Spencer, Howie, Leung, Hartmann, Möller, Morris, Shi, Feng, Hoffmann, Propping, Vasilescu, Maier, Rietschel, Zammit, Schumacher, Quinn, Schulze, Williams, Giegling, Iwata, Ikeda, Darvasi, Shifman, He, Duan, Sanders, Levinson, Gejman, Cichon, Nöthen, Gill, Corvin, Rujescu, Kirov, Owen, Buccola, Mowry, Freedman, Amin, Black, Silverman, Byerley and Cloninger2008; Schulze et al. Reference Schulze, Detera-Wadleigh, Akula, Gupta, Kassem, Steele, Pearl, Strohmaier, Breuer, Schwarz, Propping, Nöthen, Cichon, Schumacher, Rietschel and McMahon2009; Stefansson et al. Reference Stefansson, Ophoff, Steinberg, Andreassen, Cichon, Rujescu, Werge, Pietiläinen, Mors, Mortensen, Sigurdsson, Gustafsson, Nyegaard, Tuulio-Henriksson, Ingason, Hansen, Suvisaari, Lonnqvist, Paunio, Børglum, Hartmann, Fink-Jensen, Nordentoft, Hougaard, Norgaard-Pedersen, Böttcher, Olesen, Breuer, Möller, Giegling, Rasmussen, Timm, Mattheisen, Bitter, Réthelyi, Magnusdottir, Sigmundsson, Olason, Masson, Gulcher, Haraldsson, Fossdal, Thorgeirsson, Thorsteinsdottir, Ruggeri, Tosato, Franke, Strengman, Kiemeney, Melle, Djurovic, Abramova, Kaleda, Sanjuan, de Frutos, Bramon, Vassos, Fraser, Ettinger, Picchioni, Walker, Toulopoulou, Need, Ge, Yoon, Shianna, Freimer, Cantor, Murray, Kong, Golimbet, Carracedo, Arango, Costas, Jönsson, Terenius, Agartz, Petursson, Nöthen, Rietschel, Matthews, Muglia, Peltonen, St Clair, Goldstein, Stefansson and Collier2009; Rivero et al. Reference Rivero, Reif, Sanjuán, Moltó, Kittel-Schneider, Nájera, Töpner and Lesch2010; Chen et al. Reference Chen, Lee, Maher, Fanous, Chen, Zhao, Guo, van den Oord, Sullivan, Shi, Levinson, Gejman, Sanders, Duan, Owen, Craddock, O'Donovan, Blackman, Lewis, Kirov, Qin, Schwab, Wildenauer, Chowdari, Nimgaonkar, Straub, Weinberger, O'Neill, Walsh, Bronstein, Darvasi, Lencz, Malhotra, Rujescu, Giegling, Werge, Hansen, Ingason, Nöethen, Rietschel, Cichon, Djurovic, Andreassen, Cantor, Ophoff, Corvin, Morris, Gill, Pato, Pato, Macedo, Gurling, McQuillin, Pimm, Hultman, Lichtenstein, Sklar, Purcell, Scolnick, St Clair, Blackwood and Kendler2011; Cichon et al. Reference Cichon, Mühleisen, Degenhardt, Mattheisen, Miró, Strohmaier, Steffens, Meesters, Herms, Weingarten, Priebe, Haenisch, Alexander, Vollmer, Breuer, Schmäl, Tessmann, Moebus, Wichmann, Schreiber, Müller-Myhsok, Lucae, Jamain, Leboyer, Bellivier, Etain, Henry, Kahn, Heath, Hamshere, O'Donovan, Owen, Craddock, Schwarz, Vedder, Kammerer-Ciernioch, Reif, Sasse, Bauer, Hautzinger, Wright, Mitchell, Schofield, Montgomery, Medland, Gordon, Martin, Gustafsson, Andreassen, Djurovic, Sigurdsson, Steinberg, Stefansson, Stefansson, Kapur-Pojskic, Oruc, Rivas, Mayoral, Chuchalin, Babadjanova, Tiganov, Pantelejeva, Abramova, Grigoroiu-Serbanescu, Diaconu, Czerski, Hauser, Zimmer, Lathrop, Schulze, Wienker, Schumacher, Maier, Propping, Rietschel and Nöthen2011; Hansen et al. Reference Hansen, Ingason, Djurovic, Melle, Fenger, Gustafsson, Jakobsen, Rasmussen, Tosato, Rietschel, Frank, Owen, Bonetto, Suvisaari, Thygesen, Pétursson, Lönnqvist, Sigurdsson, Giegling, Craddock, O'Donovan, Ruggeri, Cichon, Ophoff, Pietiläinen, Peltonen, Nöthen, Rujescu, St Clair, Collier, Andreassen and Werge2011, Ripke et al. Reference Ripke, Sanders, Kendler, Levinson, Sklar, Holmans, Lin, Duan, Ophoff, Andreassen, Scolnick, Cichon, St Clair, Corvin, Gurling, Werge, Rujescu, Blackwood, Pato, Malhotra, Purcell, Dudbridge, Neale, Rossin, Visscher, Posthuma, Ruderfer, Fanous, Stefansson, Steinberg, Mowry, Golimbet, De Hert, Jönsson, Bitter, Pietiläinen, Collier, Tosato, Agartz, Albus, Alexander, Amdur, Amin, Bass, Bergen, Black, Børglum, Brown, Bruggeman, Buccola, Byerley, Cahn, Cantor, Carr, Catts, Choudhury, Cloninger, Cormican, Craddock, Danoy, Datta, de Haan, Demontis, Dikeos, Djurovic, Donnelly, Donohoe, Duong, Dwyer, Fink-Jensen, Freedman, Freimer, Friedl, Georgieva, Giegling, Gill, Glenthøj, Godard, Hamshere, Hansen, Hansen, Hartmann, Henskens, Hougaard, Hultman, Ingason, Jablensky, Jakobsen, Jay, Jürgens, Kahn, Keller, Kenis, Kenny, Kim, Kirov, Konnerth, Konte, Krabbendam, Krasucki, Lasseter, Laurent, Lawrence, Lencz, Lerer, Liang, Lichtenstein, Lieberman, Linszen, Lönnqvist, Loughland, MacLean, Maher, Maier, Mallet, Malloy, Mattheisen, Mattingsdal, McGhee, McGrath, McIntosh, McLean, McQuillin, Melle, Michie, Milanova, Morris, Mors, Mortensen, Moskvina, Muglia, Myin-Germeys, Nertney, Nestadt, Nielsen, Nikolov, Nordentoft, Norton, Nöthen, O'Dushlaine, Olincy, Olsen, O'Neill, Orntoft, Owen, Pantelis, Papadimitriou, Pato, Peltonen, Petursson, Pickard, Pimm, Pulver, Puri, Quested, Quinn, Rasmussen, Réthelyi, Ribble, Rietschel, Riley, Ruggeri, Schall, Schulze, Schwab, Scott, Shi, Sigurdsson, Silverman, Spencer, Stefansson, Strange, Strengman, Stroup, Suvisaari, Terenius, Thirumalai, Thygesen, Timm, Toncheva, van den Oord, van Os, van Winkel, Veldink, Walsh, Wang, Wiersma, Wildenauer, Williams, Williams, Wormley, Zammit, Sullivan, O'Donovan, Daly and Gejman2011; Steinberg et al. Reference Steinberg, de Jong, Andreassen, Werge, Børglum, Mors, Mortensen, Gustafsson, Costas, Pietiläinen, Demontis, Papiol, Huttenlocher, Mattheisen, Breuer, Vassos, Giegling, Fraser, Walker, Tuulio-Henriksson, Suvisaari, Lönnqvist, Paunio, Agartz, Melle, Djurovic, Strengman, Jürgens, Glenthøj, Terenius, Hougaard, Ørntoft, Wiuf, Didriksen, Hollegaard, Nordentoft, van Winkel, Kenis, Abramova, Kaleda, Arrojo, Sanjuán, Arango, Sperling, Rossner, Ribolsi, Magni, Siracusano, Christiansen, Kiemeney, Veldink, van den Berg, Ingason, Muglia, Murray, Nöthen, Sigurdsson, Petursson, Thorsteinsdottir, Kong, Rubino, De Hert, Réthelyi, Bitter, Jönsson, Golimbet, Carracedo, Ehrenreich, Craddock, Owen, O'Donovan, Ruggeri, Tosato, Peltonen, Ophoff, Collier, St Clair, Rietschel, Cichon, Stefansson, Rujescu and Stefansson2011; Williams et al. Reference Williams, Craddock, Russo, Hamshere, Moskvina, Dwyer, Smith, Green, Grozeva, Holmans, Owen and O'Donovan2011) (see Table 2) using the KASP™ (competitive allele-specific PCR) genotyping system. All SNPs were under Hardy–Weinberg equilibrium (p > 0.05), calculated using Fisher's exact test. The genotype of each SNP was orthogonally coded and the values for each subject collated into a vector that could be entered into a SVM. In cases where one or more SNPs could not be genotyped for a given subject, these were excluded for all other pairs in the SVM comparison since each vector length must be the same. The number of SNPs therefore entered into the SVM for FEP versus HC, UHR versus HC, and FEP versus UHR were 20, 20 and 19, respectively. In cases where subjects declined to provide a DNA sample, a reduced number of SVM subject pairs was examined.
Table 2. Specific single nucleotide polymorphisms selected as support vector machine input and corresponding publication from which they were derived

ZNF804A, Zinc finger protein 804A; CACNA1C, calcium channel, voltage dependent, L-type, alpha 1 subunit; MHC/PRSS, major histocompatibility complex/cationic trypsinogen gene; TCF4, transcription factor 4; MMP16, matrix metallopeptidase 16; NRGN, neurogranin; CMYA5, cardiomyopathy associated 5; NCAN, neurocan; PBRM1, protein polybromo1; CCDC68, coiled coil domain containing 68; AHIL, Abelson helper integration 1; PCGEM1, prostate-specific transcript 1; CNNM2, cyclin M2; NT5C2, 5′-nucleotidase cytosolic II; ANK3, ankyrin 3; CSMD1, CUB and sushi multiple domains 1; TCF7L2, transcription factor 7-like-2; VRK2, vaccinia-related kinase 2; DYPD, dihydropyrimidine dehydrogenase; TRIM26, tripartite motif containing 26.
Neuropsychology
Each subject's answers were entered into the CVLT-II software package and a summary of raw and standardized scores generated for each task component. These scores were then collated into a single vector, reflecting the subject's performance across the test, which could be entered into a SVM. Since one UHR subject did not complete the task, only 18 SVM subject pairs were examined for the UHR versus HC CVLT-II-based comparison.
SVM
Each subject's data (segmented GM images, FA skeletons, HSCT contrast images, orthogonally coded genotype data or CVLT-II score vectors) were entered separately into SVMs (Burges, Reference Burges1998) as implemented in the PROBID software package (http://www.brainmap.co.uk/probid.htm) running under Matlab7.1 (Math Works, USA) in order to assess the diagnostic potential of each modality with respect to UHR and FEP subjects relative to HCs, and also to each other. For each comparison, subject pairs matched for age (± 4 years) and gender were used to construct samples for the classifier, with each individual scan treated as a data point located in high-dimensional space and assigned by the operator to a given class. SVM comparator groups comprised 19, 19 and 15 subject pairs for FEP versus HC, UHR versus HC and FEP versus UHR, respectively. Each classifier was embedded in a leave-one-out cross-validation (LOOCV) framework, whereby all input vectors except those from one pair (one subject from each group) were used as training data for the classifier and the remaining pair withheld as test data. The accuracy of the classifier was calculated by taking the mean of its sensitivity and specificity (Hastie et al. Reference Hastie, Tibshirani and Friedman2001) across all LOOCV folds. Statistical significance of the accuracy was determined by a permutation test, whereby subjects were randomly assigned to a class and the LOOCV cycle repeated 1000 times. This provided a distribution of accuracies reflecting the null hypothesis that the classifier did not exceed chance. The number of times where it was greater than or equal to the true accuracy was then divided by 1000 to estimate a p value for the accuracy. For each neuroimaging comparison a discrimination map was produced visualizing each voxel's weight-vector score (wi) – representing its relative contribution in defining the OSH – displaying the pattern of regions able to discriminate each group. Unlike previous studies (Mourão-Miranda et al. Reference Mourão-Miranda, Bokde, Born, Hampel and Stetter2005; Marquand et al. Reference Marquand, Mourão-Miranda, Brammer, Cleare and Fu2008) no map threshold was applied since any successful discrimination was founded on the total number of voxel intensities entered into the SVM. For successful genetic and CVLT-II-based classifiers, analogous graphs showing the wi for each SNP or CVLT-II subcomponent, respectively, were also produced. Given that redundant feature extraction was not employed, nor a priori regions of interest specified, it was not possible to draw inferences regarding specific regions, SNPs or CVLT-II subcomponents out of the context of the overall pattern unlike mass-univariate results. For all classifiers, a linear kernel was used and the SVM parameter C was fixed to unity.
SVM classification accuracies
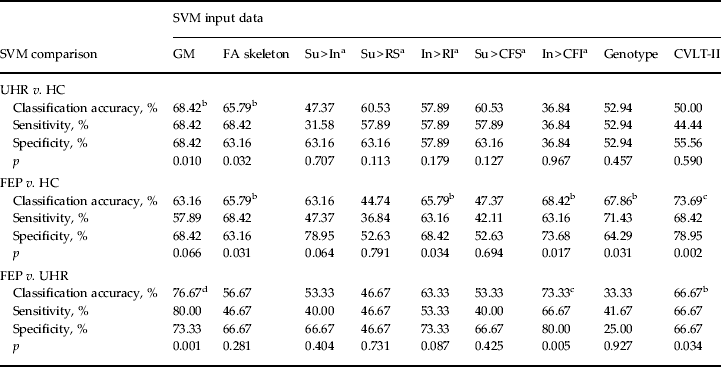
In order to correct for multiple comparisons we employed both a Holm–Bonferroni step-down procedure, which controls for family-wise error (FWE) (Holm, Reference Holm1979), in addition to the generally less conservative Benjamini–Hochberg procedure, which controls for false discovery rate (FDR) (Benjamini & Hochberg, Reference Benjamini and Hochberg1995). Since both procedures are intended for independent data, however, which the comparisons here are unlikely to be, there is an increased risk of type II error. Therefore, in the absence of an optimally established method for correcting non-independent hypotheses, for completeness both corrected and uncorrected accuracies are reported (see Table 3).
Table 3. Classification accuracy, sensitivity, specificity and p value for each binary group comparison, using sMRI, DTI, fMRI, genetic and cognitive input data

sMRI, Structural magnetic resonance imaging; DTI, diffusion tensor neuroimaging; fMRI, functional magnetic resonance imaging; SVM, support vector machine; GM, grey matter; FA skeleton, fractional anisotropy skeleton; Su, suppression; In, initiation; RS, repetition of ‘REST’ during suppression; RI, repetition of ‘REST’ during Initiation; CFS, cross-fixation during suppression; CFI, cross-fixation during initiation; CVLT-II, California Verbal Learning Test – second edition; UHR, ultra-high risk; HC, healthy control; FEP, first-episode psychosis.
a Hayling sentence completion task contrast conditions.
b p < 0.05 uncorrected.
c p < 0.05 false discovery rate-corrected.
d p < 0.05 family-wise error-corrected.
Comparing classifiers
Though the study's primary focus was to investigate whether each data type can, or cannot, successfully distinguish FEP and UHR subjects from HCs, and/or each other, for completeness, a non-parametric Cochran's Q test was also performed to examine whether the levels of accuracy for each classifier differed significantly for each diagnostic comparison. In the event of a significant result, a post hoc McNemar's test, with Bonferroni correction, was then applied to identify which specific classifiers were statistically different.
Standard univariate analysis
In order to compare the results of our multivariate approach with those of a standard univariate analysis, a paired t test comparing those subjects used for the corresponding SVM comparisons was conducted for GM images, FA skeletons, HSCT contrasts and CVLT-II scores.
Results
Demographics
There were no significant differences with respect to pre-morbid IQ between any of the groups (p > 0.05), nor was there a significant difference in PANSS scores (total, positive, negative or general) between UHR and FEP subjects (p > 0.05). With respect to medication, all FEP subjects, except one, were medicated. In comparison, all UHR subjects were medication-naive, apart from two (see Table 1).
SVM classification of GM images
Using GM images, SVM was able to successfully discriminate FEP from UHR subjects and UHR from HC subjects with accuracies of 76.67% (p < 0.05, FWE corrected) and 68.42% (p < 0.05), respectively. At a trend level only, SVM was also able to discriminate FEP subjects from HCs with an accuracy of 63.16% (p = 0.066). For the FEP versus UHR comparison the regional pattern most representative of FEP subjects was more rostrally and subcortically concentrated in comparison with the UHR group. Similarly for the UHR versus HC comparison, the regional pattern that most typified the UHR group appeared concentrated in more extreme cortical, rostral and caudal regions (Fig. 1a, b).

Fig. 1. Multivariate discrimination maps for successful structural magnetic resonance imaging (MRI)-, diffusion tensor neuroimaging (DTI)- and functional MRI-based support vector machine (SVM) classifiers. (a, b) Multivariate maps showing the pattern of grey matter regions used to discriminate: (a) first-episode psychosis (FEP) and ultra-high-risk (UHR) subjects – red indicates discrimination in favour of the FEP versus the UHR group, whilst blue indicates discrimination in favour of the UHR group versus the FEP group; (b) UHR and healthy control (HC) subjects – red indicates discrimination in favour of the UHR versus the HC group, whilst blue indicates discrimination in favour of the HC group versus the UHR group. (c, d) Multivariate maps showing the pattern of white matter regions used to discriminate: (c) UHR and HC subjects – green indicates discrimination in favour of the UHR versus the HC group, whilst yellow indicates discrimination in favour of the HC group versus the UHR group; (d) FEP and HC subjects – green indicates discrimination in favour of the FEP versus the HC group, whilst yellow indicates discrimination in favour of the HC group versus the FEP group. (e–g) Multivariate maps showing the pattern of neurofunction used to discriminate: (e) FEP and HC subjects using the initiation > repetition of ‘REST’ during initiation (In > RI) contrast – gold indicates discrimination in favour of the FEP versus the HC group, whilst turquoise indicates discrimination in favour of the HC group versus the FEP group; (f) FEP and UHR subjects using the initiation > cross fixation during initiation (In > CFI) contrast – gold indicates discrimination in favour of the FEP versus the UHR group, whilst turquoise indicates discrimination in favour of the UHR group versus the FEP group; (g) FEP and HC subjects using the In > CFI contrast – gold indicates discrimination in favour of the FEP versus the HC group, whilst turquoise indicates discrimination in favour of the HC group versus the FEP group. (a–g) Left to right, axial slices with Montreal Neurological Institute (MNI) z coordinate −28, −6, 2, 16, 32, 46, 67. The colour scale for each subfigure shows the absolute value of the weight vector score for each voxel, representing its relative contribution to the optimal separating hyperplane.
SVM classification of FA skeletons
Based on FA skeletons, SVM was able to successfully discriminate both FEP subjects from HCs, and UHR subjects from HCs with 65.79% accuracy (p < 0.05). The pattern of regions used for each classification was widely and diffusely spread, with no clear concentration of regions discernible (Fig. 1c, d). In contrast, it was not possible to directly discriminate FEP from UHR subjects using DTI with significant accuracy.
SVM classification of HSCT contrasts
Of the five contrasts tested, only two were able to make successful discriminations. Using the In > RI contrast FEP subjects were distinguishable from HCs with an accuracy of 65.79% (p < 0.05). In comparison, using the contrast In > CFI, SVM could discriminate between both FEP and UHR subjects, and also between FEP subjects and HCs, with accuracies of 73.33% (p < 0.05, FDR corrected) and 68.42% (p < 0.05), respectively. As Fig. 1e–g show, the regional pattern discriminating FEP from UHR and HC subjects was concentrated in the frontal and occipital cortices; in contrast, the pattern that most typified the UHR and HC relative to FEP subjects was widespread with greater prominence in the areas encompassing the central fissure. fMRI data were unable to distinguish UHR from HC subjects.
SVM classification using genotype
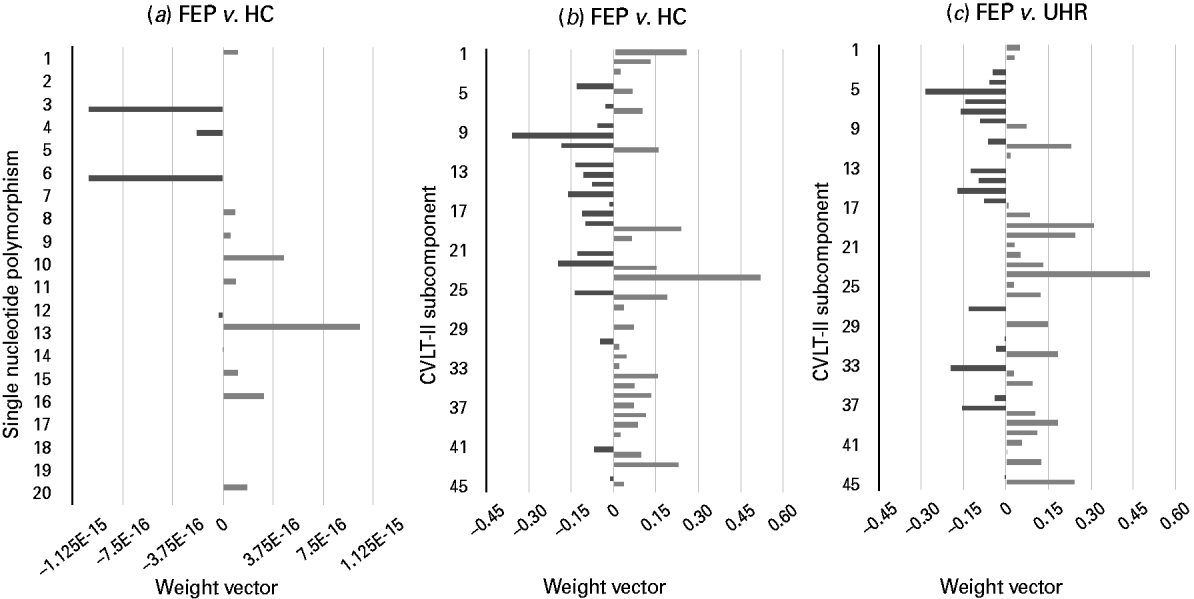
Using genetic information comprising data from a combination of 20 psychosis-associated SNPs, SVM was able to successfully discriminate FEP subjects from HCs with an accuracy of 67.86% (p < 0.05) (Fig. 2a). Comparatively, it was not possible to discriminate UHR subjects from HCs, or FEP from UHR subjects using genetic data.

Fig. 2. Weight vectors for successful genetic- and California Verbal Learning Test – second edition (CVLT-II)-based support vector machine (SVM) classifiers. (a–c). Bar charts showing the weight vector for each (a) single nucleotide polymorphism and (b, c) CVLT-II subcomponent, representing their relative contribution to the optimal separating hyperplane, used to discriminate. (a) First-episode psychosis (FEP) and healthy control (HC) subjects: light grey indicates discrimination in favour of the FEP versus the HC group, whilst dark grey indicates discrimination in favour of the HC group versus the FEP group (‘E-n’ multiplies the preceding value by (10)–n, where n is a real number). (b) FEP and HC subjects: light grey indicates discrimination in favour of the FEP versus the HC group, whilst dark grey indicates discrimination in favour of the HC group versus the FEP group. (c) FEP and ultra-high-risk (UHR) subjects: light grey indicates discrimination in favour of the FEP versus the UHR group, whilst dark grey indicates discrimination in favour of the UHR group versus the FEP group. (See Table 2 for single nucleotide polymorphisms 1–20, and Supplementary material for CVLT-II subcomponents 1–45.)
SVM classification using CVLT-II score
Based on each subject's collated score representing their performance across the CVLT-II, SVM was able to successfully discriminate both FEP from HC subjects and also FEP from UHR subjects with accuracies of 73.69% (p < 0.05, FDR corrected) and 66.67% (p < 0.05), respectively (Fig. 2b, c). In contrast, CVLT-II score could not accurately differentiate UHR from HC subjects.
Comparison of classifiers
Using a Cochran's Q test no significant differences were observed between the classifiers intended to discriminate UHR and HC subjects (Q = 8.856, p = 0.451), nor FEP and HC subjects (Q = 10.400, p = 0.319). Whilst a significant difference was observed between the classifiers intended to discriminate FEP and UHR subjects (Q = 18.353, p = 0.031), subsequent post hoc McNemar's tests (Bonferroni corrected) comparing individual classifiers were not significant (p > 0.05).
Standard univariate analysis
Paired t test analysis revealed significant (p < 0.05, FWE corrected) FA differences for the contrast HC > UHR (see Supplementary Fig. 1). Further, a number of task subcomponents of the CVLT-II differed between groups in each of the three SVM comparisons (see Supplementary Table 1). In contrast, no significant differences were detected for GM or HSCT contrast data, with respect to any of the corresponding SVM comparisons.
Potential confounds
Since most FEP subjects were medicated we examined whether any successful classifier able to discriminate them from UHR, or HC, subjects could possibly be driven by this potential confound. This was achieved by performing a Pearson's correlation analysis between the projection of each FEP subject's input data onto the weight vector (i.e. the distance of each test subject's scan from the hyperplane, quantifying the relative ease, or difficulty, with which they were categorized) and their corresponding medication measure, i.e. total dose and mean dose per day. No significant correlations were found (see Supplementary material for details).
Discussion
In agreement with our first hypothesis, FEP subjects were most readily discriminable from HCs with accurate classifiers generated by all modalities, with the exception of sMRI. Notably, this demonstrates the novel finding that genetic data may be used to discriminate FEP subjects from HCs at the individual level with significant accuracy. As proposed in our second hypothesis, we show that UHR subjects are differentiable from HCs based on sMRI and DTI; however, it was not possible to distinguish these two groups using fMRI, cognitive or genetic data. Finally, consistent with our third hypothesis, we found that FEP and UHR subjects could be accurately differentiated when compared directly using sMRI, fMRI and cognitive, but not genetic or DTI, data.
Taken together, the results provide a number of possible inferences associated with the UHR and FEP states. Given that DTI data were able to successfully discriminate between the two patient groups relative to HCs, for example, supports the notion that WM alterations are associated with psychosis risk. Likewise, the fact that sMRI was able to discriminate UHR from FEP and HC subjects may imply that GM alterations are specifically associated with the UHR state, but not those who have transitioned. The absence of GM alteration in FEP subjects relative to HCs is surprising, however, given previous reports of widespread significant effects (Shepherd et al. Reference Shepherd, Laurens, Matheson, Carr and Green2012). One speculative explanation for this is that the FEP subjects recruited here were less clinically severe than those of previous studies, potentially resulting in less severe alteration. This is supported by their relatively stable symptom profile (Table 1), which may possibly, in turn, have been driven by downstream effects of exposure to anti-psychotic medication. This interpretation is made with caution, however, since the precise effects of such exposure remain unclear (Navari & Dazzan, Reference Navari and Dazzan2009), and since a successful classifier was not generated furthermore, this could also not be investigated quantitatively using a correlation analysis. In comparison, the fact that fMRI and cognitive data could differentiate FEP from UHR and HC subjects suggests that alterations in these two domains are specifically associated with conversion to psychosis. This inference is also true for individual genotype data, which were able to discriminate only between those with a FEP and HCs.
Methodologically, our results consolidate the notion that multivariate techniques such as SVM may be better suited to the development of a real-world clinical diagnostic tool than standard mass-univariate methods. Although no focal abnormalities survived univariate threshold either for sMRI nor fMRI, for example, overall patterns of alteration in data from these two modalities were still able to successfully discriminate between subjects. Furthermore, the ability to accurately distinguish FEP from HC subjects using genetic data supports the notion that individuals who suffer a FEP may be genetically predisposed to transitioning (Kéri et al. Reference Kéri, Kiss and Kelemen2009). As a non-invasive, easily obtained and relatively cheap data type, it could potentially serve as a good basis for future diagnostic tools in conjunction with clinical assessment. Similarly, the fact that CVLT-II score was able to distinguish FEP from both UHR and HC subjects may represent another non-invasive and inexpensive tool to inform identification of individuals with a FEP. With specific regard to UHR subjects in comparison, the fact that only sMRI and DTI were able to distinguish them from HCs might suggest that they are associated with patterns of neuroanatomical alteration that may occur in the absence of similar genetic, neurofunctional or cognitive patterns of alteration, though focal abnormalities may still be evident. This aspect of our results therefore provides tentative support for the use of sMRI and DTI as a clinical aid in identifying those at UHR of psychosis, but may be limited by the associated costs and technical expertise involved.
It should be acknowledged, however, that in comparison with the few previous studies to have applied SVM to the UHR cohort, the accuracies found here discriminating UHR from HC subjects were relatively modest (Koutsouleris et al. Reference Koutsouleris, Meisenzahl, Davatzikos, Bottlender, Frodl, Scheuerecker, Schmitt, Zetzsche, Decker, Reiser, Möller and Gaser2009a, Reference Koutsouleris, Davatzikos, Bottlender, Patschurek-Kliche, Scheuerecker, Decker, Gaser, Möller and Meisenzahl2011). As with many univariate studies, this inconsistency may have arisen from a number of possible methodological differences which include, but are not restricted to, the assessment tools used to identify subjects at UHR; the strength of the scanner and the acquisition sequence used for the collection of neuroimaging data; the data-processing pipeline used to construct features for input into SVM; and the choice of SVM parameter settings (Caruana & Niculescu-Mizil, Reference Caruana and Niculescu-Mizil2006; Orrù et al. Reference Orrù, Pettersson-Yeo, Marquand, Sartori and Mechelli2012). Indeed, it is perhaps worth noting that as a relatively novel application to the field of psychiatry, efforts to identify the optimal criteria necessary for accurate discrimination using SVM are currently ongoing, of which these studies represent some examples.
In the context of developing real-world diagnostic tools, therefore, two notes of caution must be considered. First, the eventual use of genetic, neuropsychological and multimodal neuroimaging data in clinical practice would arguably require greater levels of diagnostic accuracy than those found here. One avenue to achieving this may lie in the integration of different types of data within the same SVM allowing information from one modality to inform that of another, for example, as used recently by Yang et al. (Reference Yang, Liu, Sui, Pearlson and Calhoun2010) to discriminate ChSz patients from HCs. It remains, however, that any future translational implementation of SVM must account for the fact that the impact of misclassifying someone ill as healthy may be worse than misclassifying someone healthy as ill. As such, a classifier able to detect patients with excellent sensitivity, but healthy individuals with only good specificity, may be preferred to a classifier with excellent specificity but only good sensitivity. Second, it should be noticed that the application of SVM could only reach the same level of diagnostic accuracy as traditional methods of clinical assessment since the development of the classifier is based on the distinction between groups in the training data, which ultimately relies on traditional diagnostic methods. Nevertheless, such technology may help in a clinical setting by discriminating between those most difficult to categorize using traditional methods of assessment alone. Furthermore, it could potentially be used in a forensic setting as an objective means of reducing controversy in evaluations of mental illness and minimizing errors in detecting malingering (Sartori et al. Reference Sartori, Pellegrini and Mechelli2011).
Limitations
The study's primary limitation was that at the time of scanning, the majority of FEP patients were medicated, and correspondingly, symptomatically stable. Although we found no evidence for a significant impact of medication, it is possible that anti-psychotic exposure, or even other variables not considered here, may still have contributed to the classification in an as yet undetectable way, potentially confounding the inference one can draw from the successful discriminator. It should also be acknowledged furthermore that the medication measures used as variables to detect potential confounds (i.e. total dose, average dose) may not have fully captured the historical and cumulative effects of exposure to anti-psychotics which may, in comparison, be more severe. Consequently, it cannot be ruled out, for example, that the successful FEP classifiers were simply distinguishing subjects who have, or have not, been exposed to anti-psychotics. However, it remains that since the exact nature and extent of the effects of anti-psychotic medication on brain structure are not yet known (Navari & Dazzan, Reference Navari and Dazzan2009), this is an issue not specific to the current study, but is instead one that applies to the vast majority of studies of psychiatric patients. A second limitation is that, in the absence of an optimally established method for correcting for non-independent comparisons, we used two types of correction intended for multiple independent comparisons which may have resulted in an increased risk of type II error. However, we also reported uncorrected results for completeness. A third limitation, applicable to any study with access restricted to their own sample, is that as a single-centre, cross-sectional, study we are unable to make inference regarding the generalizability across different research centres for any of the successful classifiers, nor at this stage make any prediction of subsequent progression within the UHR group. A fourth limitation of the study is that the three subject groups were not compared using a multi-class classification approach, which would have provided a closer approximation of how differential diagnostic decisions are made in real-world clinical practice. However, since multiple binary classifiers for each possible comparison were generated in the current study, a quantitative estimate is still provided demonstrating the relative ease, or difficulty, with which subjects from each group may be differentiated from the subjects of every other group, with respect to each data type. Lastly, since we did not use any a priori regions of interest or automated data-driven feature selection, we are unable to make inferences regarding specific neuroanatomical regions, CVLT-II task components, SNPs, or risk alleles, since in each case the entirety of the data entered into the SVM was used to generate the classifier.
Conclusion
The evidence presented here demonstrates that subjects who have had a FEP can be identified at the individual level using a range of biological and cognitive measures including genetic, DTI, fMRI and cognitive data. In contrast, sMRI and DTI were the only modalities that allowed identification of those at UHR of psychosis with significant accuracy. For the first time we have shown that FEP and UHR subjects can be directly differentiated using neuropsychological, sMRI and fMRI data. From a clinical perspective, the results provide preliminary support to the translational development of SVM as a clinically useful diagnostic aid, highlighting patterns of genetic, cognitive, neuroanatomical and neurofunctional alterations that could, in the future, be potentially used to inform identification of those with subclinical symptomatology and recent converters. Nevertheless, we would stress that the eventual use of this approach in everyday clinical practice would arguably require greater levels of diagnostic accuracy than found in the present study, with the integration of data representing one possible solution.
Supplementary material
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S003329171300024X.
Acknowledgements
This work was supported by the Wellcome Trust (no. WT085390/Z/08/Z). W.P.-Y. was supported by a Ph.D. studentship from the Medical Research Council. A.F.M. gratefully acknowledges support from King's College Annual Fund and the King's College London Centre of Excellence in Medical Engineering, funded by a Wellcome Trust and Engineering and Physical Sciences Research Council (EPSRC) grant (no. WT088641/Z/09/Z). D.P. was supported by a National Institute for Health Research (NIHR) post-doctoral fellowship. A.M. was supported by an Investigator Award from the National Alliance for Research on Schizophrenia and Depression (NARSAD). The authors thank the NIHR Biomedical Research Centre for Mental Health at the South London and Maudsley NHS Foundation Trust for its continued support of our translational research objectives.
Declaration of Interest
None.