Ultrasound transducers that contact broken skin or mucous membranes are considered semicritical devices and should undergo high-level disinfection (HLD) between patients.Reference Rutala and Weber 1 Studies have shown residual contamination on ultrasound transducers when HLD is not performedReference Casalegno, Le Bail Carval and Eibach 2 , Reference Leroy 3 and cases of hepatitis B and C infection have been reported where transmission was thought to have been caused by improper reprocessing of ultrasound transducers.Reference Ferhi, Roupret, Mozer, Ploussard, Haertig and de La Taille 4 , 5
HLD methods that involve soaking ultrasound transducers in liquid chemicals, such as glutaraldehyde or ortho-phthalaldehyde, do not typically disinfect the handle. Some major manufacturers specifically advise against submersion of the handle owing to the transducer not being fully sealed. There are no clear recommendations on whether transducer handles should be disinfected, nor is there any consensus on the risk that contaminated transducer handles might pose to patients.
Although full immersion of transducers in liquid is not generally possible, approaches that allow the HLD of the entire transducer are now available. This study surveyed the residual contamination on transducer handles when not disinfected (routine soaking method) and also assessed the efficacy of an automated disinfection device capable of disinfecting both heads and handles of ultrasound transducers.
METHODS
This study was a prospective, bi-centric, cross-sectional study, performed at an ultrasound clinic and public hospital. No patients were excluded and no patient data were collected so the study did not require ethics approval. Following routine ultrasound procedures, all transducers were cleaned with detergent and water and were dried with paper towels. Handle samples were then collected from 2 study groups after the handles were subjected to different disinfection methods. Groups were sampled sequentially in randomized order.
The first group of transducers (hereafter referred to as “glutaraldehyde disinfection of head only” [GDHO]) underwent reprocessing according to the existing protocol at the clinic where the head of the transducer was soaked in a 2.4% solution of glutaraldehyde for 20 minutes at room temperature. Transducer handles were not disinfected (per normal practice).
The second group of transducers (hereafter referred to as “automated disinfection of head and handle” [ADHH]) were subjected to HLD with an automated disinfection system that disinfects the entire transducer including the handle (trophon EPR; Nanosonics). The device was operated according to the manufacturer’s instructions for use.
Matched control samples (shams) were collected to quantify contamination occurring during sample collection. Positive and negative controls were included on all test days and all microbiologic testing was performed blinded.
Samples were collected immediately following reprocessing by swabbing transducer handles with 2 sterile cotton swabs dipped in tryptone soya broth (TSB). Gloves were worn and care was taken not to contaminate the transducers. Sham samples were collected by swabbing the inside of a sterile Petri dish lid with 2 cotton swabs wetted with TSB. Swabs were snapped off into 10 mL of TSB supplemented with 0.02% catalase. Samples were stored at 4°C and were cultured within 24 hours of collection.
Each broth tube was vortexed for 60 seconds and serially diluted 1/10 with 1 mL being transferred to tryptone soya agar plates for counts with sensitivity down to 10 colony-forming units (CFU)/mL. Plates were incubated in air at 37°C for 72 h and the remaining TSB was incubated for 48 h at 37°C in air with growth indicating less than 10 CFU/mL. The predominant organism was recovered from plates or broth as appropriate and was identified to species level where possible using a VITEK device (bioMérieux). The methicillin sensitivity of Staphylococcus aureus and vancomycin sensitivity of Enterococcus faecium isolates were established using the calibrated dichotomous sensitivity (CDS) method.Reference Bell, Pham and Fisher 6
Residual handle contamination was compared across disinfection methods using χ2 and nonparametric Kruskal-Wallis tests. The contamination rate (matched sham samples) was compared with the ADHH contamination rate as binary and continuous variables using matched McNemar χ2 and Wilcoxon signed rank sum tests and visualized graphically with a bar chart and a dot plot, respectively. A generalized linear mixed model was derived to estimate the risk of contamination with each disinfection method. The modeling took into account adjustment on the contamination rate in the matched sham, clustered to the sampling day. The outcome measure was bacterial growth assessed as a binary variable (positive if>0 CFU/mL), and the risk of contamination remaining on the transducer handle was expressed as an adjusted odds ratio. A similar analysis was performed on the basis of the continuous variable of bacterial growth (CFU/mL) using a mixed linear model. This provided a regression coefficient related to the disinfection method, adjusted on the matched rate of contamination, and again clustered to the days of sampling. For the purposes of this analysis, any count that was less than 10 CFU/mL was rounded to 10 CFU/mL. No outlier was found to influence the convergence modeling. Statistical analyses used Stata/SE, version 12 (StataCorp).
RESULTS
In total, 77 and 75 handle samples were analyzed from GDHO and ADHH groups, respectively. The matched sham samples showed only small amounts of contamination (6.7% matched to the ADHH group and 9.1% matched to the GDHO group), suggesting that the sampling technique used was robust. Samples from the ADHH group showed less contamination than the matched sham samples (5.3 % vs 6.7%, P=.5 [pairwise χ2]; Figure 1A). When considered as a continuous variable, no statistically significant difference was observed between matched sham samples and ADHH samples (median, 0 [interquartile range, 0–5] for sham samples vs 0 [0–5] for ADHH samples; P=.7).
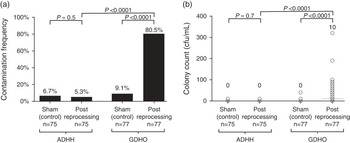
FIGURE 1 Transducer handle contamination following reprocessing. A, Samples were collected from handles following glutaraldehyde disinfection of head only (GDHO) or following automated disinfection of head and handle (ADHH) and were cultured. Corresponding contamination frequencies were compared using a pairwise χ2 test for matched samples and a χ2 test when comparing GDHO and ADHH samples. Values above each bar indicate contamination frequency. B, The extent of transducer handle contamination was determined by performing plate counts on transducer handle samples to determine the number of organisms contaminating the handle. Individual counts are represented by circles. Horizontal bars represent the median, which is given above each group. Distributions according the study group were compared using a nonparametric Kruskal-Wallis test. CFU, colony-forming units.
A large and significant difference was observed in the number of handle samples showing contamination from the GDHO versus the ADHH group (80.5% vs 5.3%; P<.0001). Bacterial contamination measured as a continuous variable was also significantly lower in the ADHH group compared with the GDHO group: median of 0 CFU/mL (interquartile range, 0 to 5) versus 10 CFU/mL (0 to 90); P=.0001 (Figure 1B).
The same results were observed when adjusting for the matched control samples, and when clustering by sampling day. The risk of contamination remaining was much higher with the GDHO group handles compared with the ADHH group (adjusted odds ratio, 82.0 [95% CI, 25.2–266.5], P<.0001). Bacterial contamination considered as a continuous variable was also significantly higher in the GDHO group compared with the ADHH group after adjusting for the control samples and sampling day (coefficient, 25.4 [95% CI, 14.9–36.1], P<.0001).
The array of organisms isolated from all samples is shown in Table 1. Of the 12 S. aureus isolates from the GDHO group, one was shown to be methicillin (cefoxitin) resistant. The E. faecium isolate from the GDHO group was sensitive to vancomycin.
TABLE 1 Isolates Recovered From Sham Samples and Ultrasound Transducer Handles Immediately Following Reprocessing
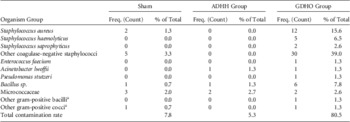
NOTE. ADHH, automated disinfection of head and handle; GDHO, glutaraldehyde disinfection of head only.
a These organisms could not be fully identified to species level.
DISCUSSION
To our knowledge, residual contamination on ultrasound transducer handles has not previously been systematically investigated. Use of glutaraldehyde-based or ortho-phthalaldehyde–based soaking methods for HLD of intracavity ultrasound transducers is common; however, these methods do not typically disinfect the handle. This study investigated the extent of handle contamination following HLD with glutaraldehyde soaking where the handle was not disinfected or following disinfection with an automated HLD system.
The inclusion of sham samples in this study enabled the estimation of contamination that occurred either during sampling or during the microbiologic procedures. Handle samples from the ADHH group showed a lower contamination rate than the sham controls, suggesting that the disinfection system was effective within the limitations of the testing methods utilized. On this basis, residual microbes found in the ADHH group are likely to have arisen from sampling contamination. The efficacy of the device is consistent with previously published laboratory-based test data showing that the device achieves HLD.Reference Vickery, Gorgis, Burdach and Patel 7
The majority (80.5%) of samples taken from the GDHO group were contaminated, with some isolates being known etiologic agents of healthcare-associated infections. In particular, a large proportion of isolates were S. aureus (15.4% of GDHO samples) and one of these isolates was found to be methicillin-resistant S. aureus (MRSA). Patients indicated for transvaginal ultrasound may include presurgical patients such as pregnant women, many of whom will subsequently undergo caesarean delivery (31.3% in the United States).Reference Osterman and Martin 8 MRSA colonization of these patients is a concern owing to the well-established link between MRSA colonization and resultant MRSA surgical wound infections.Reference Guyot and Layer 9
Causative agents of urinary tract infections including S. aureus, Staphylococcus haemolyticus, Staphylococcus saprophyticus, and E. faecium were isolated from the GDHO group. Rectal, vaginal, and urethral colonization is a risk factor for urinary tract infections and artificial introduction of these organisms into the vagina (or rectum for transrectal ultrasound) could result in colonization of these sites. Treated and untreated urinary tract infections in pregnancy can be associated with negative outcomes for both mother and baby, highlighting the need to focus on prevention in the first instance.
Presumably organisms detected on the handle may be sourced from patients, staff, or the environment, resulting in an extensive range of potential contaminants. In particular, staff handling of the handle when not disinfected could lead to contamination. This study was limited both by patient numbers (meaning that more-rare microorganisms may not have been isolated) and by the detection methods used. The culture media (TSB and tryptone soya agar) were not suitable for the detection of fastidious bacteria or fungi and the methods used were not capable of detecting viruses or parasites. Given that patients in this study were undergoing transvaginal ultrasound, significant pathogens that could conceivably contaminate handles might include fastidious bacteria such as group B streptococci and Streptococcus pyogenes, Neisseria gonorrhea, and Treponema pallidum. Moreover, the methods used would not have been able to detect sexually transmitted viral pathogens such as herpes simplex virus, human papillomavirus, human immunodeficiency virus, or the parasite Trichomonas vaginalis. A recent study has also shown that native human papillomavirus 16 is highly resistant to glutaraldehyde and ortho-phthalaldehyde.Reference Meyers, Ryndock, Conway, Meyers and Robison 10 Given that 80.5% of post-glutaraldehyde handle samples were contaminated with nonfastidious organisms, it seems likely that fastidious pathogens, viruses, or parasites might also be isolated if a sufficiently large sample size and methods appropriate for detecting these organisms were used. In addition to the automated device tested here, other disinfection methods may also be suitable for disinfecting transducer handles. Further research is required to evaluate these other methods.
This study demonstrates that ultrasound transducer handles may become contaminated with clinically significant organisms if transducers are reprocessed with a method that does not disinfect the handle. Infection control staff should consider using HLD methods that also disinfect the entire transducer.
Acknowledgments
Financial support. Nanosonics.
Potential conflicts of interest. D.P. reports that she is chief microbiologist at Nanosonics and V.G., S.L., and J.B. report that they are consultants to Nanosonics. All other authors report no conflicts of interest relevant to this article.




