INTRODUCTION
Human bocavirus (HBoV) was discovered during a meta-genomic study of pooled specimens of respiratory secretions from Swedish children with acute respiratory symptoms [Reference Allander1]. HBoV was classified in the family Parvoviridae, subfamily Parvovirinae, genus Bocavirus. It is a non-enveloped, icosahedral virus, with a 5·3 kb single-stranded DNA genome with three open reading frames [Reference Allander1]. Recent studies have revealed that there are four species of HBoV, numbered sequentially HBoV-1 to HBoV-4 [Reference Arthur2–Reference Kapoor4].
HBoV has been detected in samples from patients with symptoms of acute respiratory infection, gastroenteritis and tonsillitis in several regions of world [Reference Fry5–Reference Vicente8]. However, the association of HBoV with human respiratory and gastrointestinal infections is unclear. Similar to other viruses of the family Parvoviridae, HBoV may persist [Reference Martin9, Reference Proenca-Modena10], leading to prolonged detection of its genome in secretions. Therefore, it is unsurprising that HBoV is frequently detected simultaneously with genomes of other agents, including respiratory and enteric viruses [Reference Proenca-Modena10, Reference Bonzel11]. Detection of direct or surrogate viral replication markers in association with acute symptoms could partially help to circumvent the non-fulfilment of Koch's postulates by the lack of an experimental animal model. Recent studies have shown that only a small portion of patients with HBoV-1 and respiratory diseases have an acute infection by this virus, characterized by the presence of replicative markers, high viral load and absence of viral co-infections in nasopharyngeal secretions, in addition to the frequent simultaneous presence of diarrhoea [Reference Proenca-Modena10]. In Latin America, few studies have been conducted on the role of HBoV and gastroenteritis in children [Reference Albuquerque12, Reference Santos13]. This report describes a study of HBoV-1 viral loads, as an indicator of acute infection, in stool samples from patients with non-bacterial acute diarrhoea with or without the simultaneous detection of other enteric viruses.
METHODS
Samples
In this retrospective, cross-sectional study, 349 stool samples from children (52·5% boys) were screened by quantitative polymerase chain reaction (qPCR) for HBoV DNA. Samples were collected between January and December 2004 from children aged <5 years with acute gastroenteritis but negative for common enterobacteria by microbiological assays (Salmonella spp., Shigella spp., and enteropathogenic and enteroinvasive Escherichia coli), and without respiratory symptoms, admitted to one of the main medical centres (Sanatorio San Roque) in Paraguay [Reference Parra14, Reference Amarilla15]. Of the tested samples, 253 (72·3%) were from children aged <2 years. All samples had been previously tested for rotavirus (RV) and norovirus (NoV) [Reference Parra14, Reference Amarilla15]. All stools samples were stored at −20°C until DNA extraction. Only one clinical specimen was tested for each patient included in this study.
Ethical statement
The study was conducted according to the principles expressed in the Declaration of Helsinki and was approved by the Ethics Review Committee of Instituto de Investigaciones en Ciencias de la Salud, Universidad Nacional de Asunción (IICS-UNA), Number P45/07, and informed consent was obtained from all parents and guardians prior to enrolment.
qPCR
DNA was extracted from 300 μl of a stool suspension prepared in PBS and centrifuged at 3500 g, using a commercial kit (Wizard Genomic DNA Purification kit; Promega, USA). The qPCR assay for HBoV detection and quantification, targeted to the viral NP1 gene, was based on previously published procedures [Reference Neske16]. Amplification of serial decimal dilutions of plasmid pGEM-FULL4HBoV containing a 1 kb insert from the HBoV NP1 gene (2270–3280 nt) was included in each batch to calculate viral loads [Reference Proenca-Modena10]. The assay detection limit was 4·6 copies of HBoV plasmid, and the slope and correlation coefficient of the standard curve were −3·386 and 0·994, respectively. All applicable measures were taken to prevent cross-contamination of samples, including the use of separate rooms for DNA extraction and PCR mix preparation. Negative and positives control samples were included in all batches. All samples were tested by qPCR for β-actin as an endogenous control of sample integrity, as described previously [Reference Proenca-Modena10].
Phylogenetic analysis
All HBoV-positive samples were tested by PCR to obtain a 500 bp product from the HBoV VP1/VP2 region (nt 3640–4140) using primers VP1-F (5′-ACAATTCAGAATGGTCACCTCTACA-3′) and VP2-R (5′-TTGGTGGTCACTGTTTTCAAG-3′). All obtained products were excised from 1·0% agarose gels, purified with QIAquick Gel Extraction kit (Qiagen, Germany) and sequenced with BigDye Terminator v. 3.1 Cycle Sequencing (Applied Biosystems, USA) using the ABI Prism 3500 Genetic Analyzer. Both DNA strands were sequenced using VP1-F and VP2-R primers. A representative Paraguayan isolate, Py380SR04, was deposited in the GenBank database (accession no. JN701022). The 37 sequences obtained were manually edited using Bioedit v. 7.0.9 [Reference Hall17] and phylogenetic analysis was performed with an additional 45 sequences of HBoV (1–4) VP1/VP2 genes retrieved from GenBank, aligned with Clustal X 5.2 [Reference Larkin18], using MEGA v. 5.0 [Reference Tamura19], with Kimura-2 as the substitution model for nucleotide, and statistically supported by the bootstrap method using 1000 replicates.
RESULTS
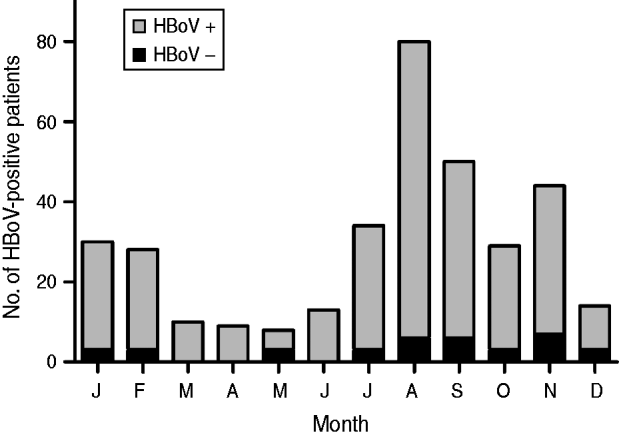
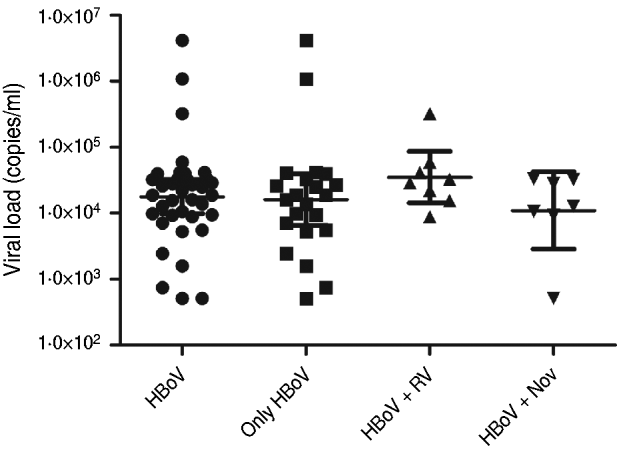
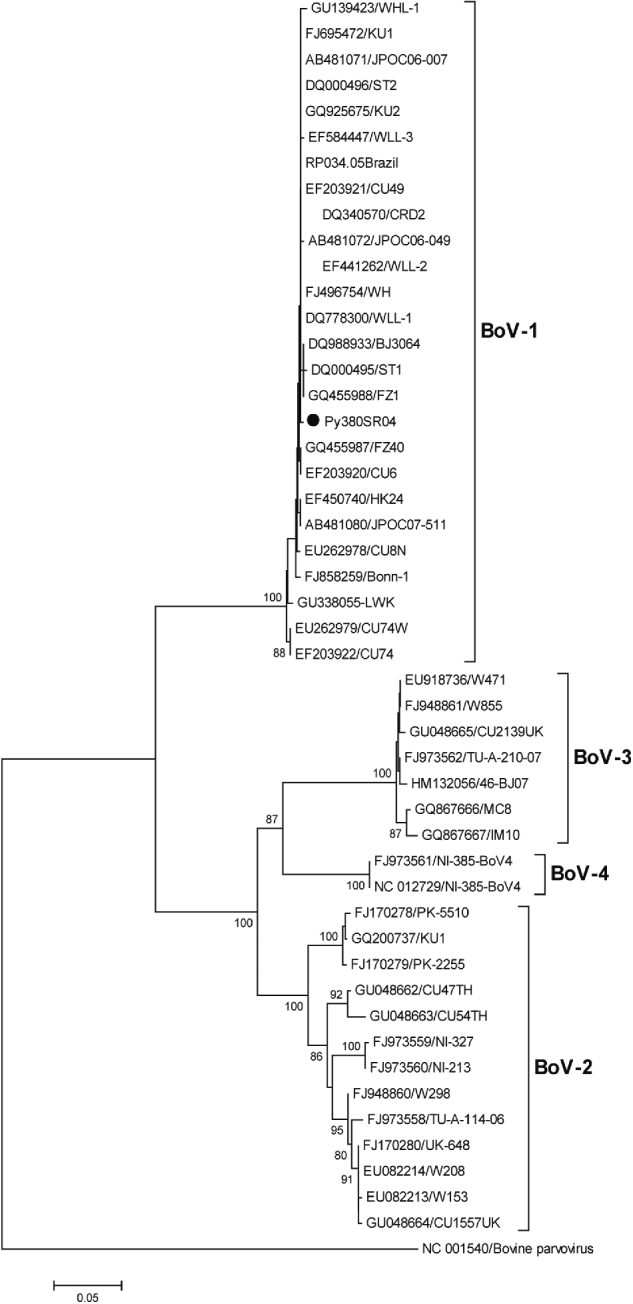
HBoV-1 genome was detected by qPCR in 37 (10·6%) samples, the majority (67·6%) of which were from children aged <2 years (Table 1). No significant association was noted between the presence of HBoV-1 and child age or gender. Monthly distribution showed no clear seasonality (Fig. 1). Co-infection with RV or NoV was detected in 15 (40·5%) of the 37 HBoV-positive samples. RV was detected in eight (21·6%) and NoV in seven (18·9%), and co-detection of RV and NoV was not found (Table 1). Absolute quantification of HBoV DNA by qPCR revealed broad variation in samples, with a median 1·88 × 104 (mean ± s.d.: 1·69 × 105 ± 6·9 × 105) copies/ml (Fig. 2). The median viral load in stools in which only HBoV-1 was detected was 1·74 × 104 (2·54 × 10 ± 9·02 × 105) copies/ml, whereas in samples from patients with HBoV-1 and RV co-infection it was 3·07 × 104 (6·64 × 104 ± 1·05 × 105) copies/ml, and in patients with HBoV-1 and NoV co-infection it was 1·27 × 104 (1·81 × 104 ± 1·29 × 104) copies/ml (Fig. 2). There was no association of high viral load with any pattern of viral co-infections. The sequencing of 500 bp in the VP1/VP2 region of HBoV genomes confirmed they were all HBoV-1 (Fig. 3), and revealed ∼99% sequence similarity (data not shown).

Fig. 1. Monthly distribution of HBoV-positive stool samples from children with acute gastroenteritis in Paraguay, 2004.

Fig. 2. Viral loads of HBoV-1 (copies of genome/ml) determined by quantitative polymerase chain reaction in stool samples from patients with HBoV-1 as single agent, and with simultaneous detection of rotavirus (RV) and norovirus (NoV).

Fig. 3. Phylogenetic tree constructed with nucleotide sequences of 46 HBoVs in a region of the VP1/VP2 gene (500 bp). Because of the high sequence similarity of all 37 Paraguayan strains (>99%), only one representative strain (Py380SR04) is shown in the tree (solid black circle). The outgroup sequence corresponds to a strain of bovine parvovirus. Bootstrap values <70% are not shown.
Table 1. Age and gender distribution of positive samples for HBoV-1 alone or in association with other viruses

RV, Rotavirus; NoV, norovirus.
DISCUSSION
HBoV-1 has been detected in studies conducted around the world, usually in association with respiratory infections [Reference Allander20]. HBoV detection in stool samples from patients with acute gastroenteritis has been reported to range from 2% to 20% [Reference Cheng21–Reference Nadji24]. In the present study, about 10% of the tested samples were positive for HBoV-1 DNA by qPCR, which was confirmed by an independent amplification of another genomic region with sequence analysis of the VP1 gene. In a study recently conducted in Brazil [Reference Albuquerque12], a frequency of 2% of HBoV was reported in children aged <15 years with acute diarrhoea between 2003 and 2005. The difference in frequencies between these two studies probably reflects variations in yearly rates of HBoV circulation, and the ages of patients. In fact, studies have shown that HBoV can circulate with different rates in consecutive years [Reference Maggi25], and is more prevalent in younger patients [Reference Karalar26]. Moreover, exclusion of patients with a positive stool sample for common enterobacteria may have contributed to the higher HBoV frequency observed.
The role of HBoV-1 as causative agent of gastroenteritis in humans is still unclear and it is reasonable to assume that HBoV-1 is in fact a virus of the respiratory tract, which is detected in stools as a remnant of its passage through the gastrointestinal tract. Despite the fact that in ∼60% of the HBoV-1-positive samples this agent was the only virus detected, this is not sufficient to support its role in the aetiology of gastroenteritis. Therefore, determination of HBoV-1 viral load was attempted to further substantiate this issue, bearing in mind previous results obtained with HBoV-2, a predominantly enteric HBoV that may reach up to 109 DNA copies/ml of stools [Reference Kantola27]. The present analysis showed that viral loads varied broadly, with no correlation with detection of HBoV-1 as the only detectable virus in stools. This is in contrast with a previous study conducted in patients with respiratory diseases, which showed that HBoV viral loads can be as high as 1012 copies of viral genome/ml of nasopharyngeal secretions, and that higher viral loads correlated with lack of simultaneous detection of other respiratory viruses [Reference Proenca-Modena10].
The present results support the hypothesis that HBoV-1 replicates primarily in the respiratory tract, and that swallowed nasopharyngeal secretions might serve as a source of virus contamination of the stools. This hypothesis is not contradicted by the fact that the stools tested in the present study were from children with diarrhoea without respiratory illnesses, as asymptomatic prolonged shedding of respiratory HBoV is frequent [Reference Martin9]. In addition, there is evidence of HBoV persistence in respiratory and gastrointestinal tracts. HBoV-1 is highly frequent in adenoids and palatine tonsils of patients with adenotonsillar chronic diseases without respiratory symptoms [Reference Proenca-Modena28] and the episomal genomes of HBoV-1, -2 and -3 were detected in clinical samples from humans [Reference Lusebrink29–Reference Zhao31].
Given their propensity for prolonged asymptomatic shedding, definitive association between HBoVs and gastroenteritis awaits fulfilment of classic or alternative Koch's postulates. Before that becomes achievable by in vivo experimentation in animal models, prospective studies of clinical samples, with simultaneous sampling of respiratory secretions, stools and blood, and detection of HBoV replication markers are still needed. Such studies should ideally be supported by in situ methods to demonstrate HBoV replication in gastrointestinal mucosa either in vivo or ex vivo.
Although multiple viruses and bacteria have been associated with gastroenteritis, consistently high proportions of diarrhoeal diseases remain without identification of a pathogen. Therefore, the continuous discovery of new agents of diarrhoeal diseases, especially based on meta-genomic approaches, will hopefully close this gap and eventually help the development of effective interventions to lessen the burden of this disease.
ACKNOWLEDGEMENTS
This research was partially funded by Sustainable Science Institute (G.I.P.), FAPESP and CNPq (J.L.P.M, V.H.A, E.A.). The funders had no role in design, data collection, analysis, or interpretation of the data.
DECLARATION OF INTEREST
None.