Introduction
Theobroma cacao L. is the most economically important species of the genus Theobroma, a member of the Malvaceae family. Cacao fruits provide the raw material for a multibillion-dollar chocolate industry and other cocoa-based products processing pharmaceutical and cosmetic industries (Wickramasuriya and Dunwell, Reference Wickramasuriya and Dunwell2018). Cocoa demand is expected to grow at 7.3% from 2019 to 2025 to reach a market value of US$16.3 billion, due to increasing demand from emerging economies and sustained demand from developed economies (Voora et al., Reference Voora, Larrea, Huppé and Nugnes2022). Most of the world's cocoa production (80–90%) comes from 40 million smallholder farmers (up to five hectares), which is their primary source of revenue; therefore, this crop helps in the alleviation of poverty in cocoa-producing regions (Beg et al., Reference Beg, Ahmad, Jan and Bashir2017; Benjamin et al., Reference Benjamin, Lundy, Abbott, Burniske, Croft, Fenton, Kelly, Rodriguez-Camayo and Wilcox2018). The demand for fine or flavour chocolate has increased (Fernández-Niño et al., Reference Fernández-Niño, Rodríguez-Cubillos, Herrera-Rocha, Anzola, Cepeda-Hernández, Aguirre Mejía, Chica, Olarte, Rodríguez-López, Calderón, Ramírez-Rojas, Del Portillo, Restrepo and González Barrios2021), its market has been growing at a rate of 7–11% per year since 2011 (Vignati and Gómez-García, Reference Vignati and Gómez-García2020) and Colombia is one of the known producers of this kind of cocoa (Ballesteros et al., Reference Ballesteros, Lagos and L2016); however, its production is small compared to the primary producer's countries (FAO, 2022).
The increasing human population, the destruction of the Amazonian rainforests, the loss of traditional varieties and climate change have significantly altered the cacao diversity causing its vulnerability to sudden changes in weather and the appearance of new pests and diseases (CacaoNet, Reference Laliberté2012; Cilas and Bastide, Reference Cilas and Bastide2020). Cacao breeders look to improve yield, disease resistance and bean quality traits to support the growing global cacao industry (Bekele and Phillips-Mora, Reference Bekele, Phillips-Mora, Al-Khayri, Jain and Johnson2019; Rodriguez-Medina et al., Reference Rodriguez-Medina, Caicedo Arana, Sounigo, Argout, Alvarado and Yockteng2019). However, this breeding is limited by long-generation cycles, self-incompatibility, pollination inefficiency, challenging abiotic and biotic stress factors, including several major diseases (Bekele and Phillips-Mora, Reference Bekele, Phillips-Mora, Al-Khayri, Jain and Johnson2019), such as Frosty pod rot caused by Moniliophthora roreri and witches' broom caused by Moniliophthora perniciosa (Álvarez et al., Reference Álvarez, Martínez and Coy2014; Díaz-Valderrama et al., Reference Díaz-Valderrama, Leiva-Espinoza and Catherine Aime2020).
Plant breeders rely on crop genetic resources conserved in germplasm collections to incorporate genetic diversity into commercialized cultivars and develop new materials for the sustainable cultivation of cacao (Zhang and Motilal, Reference Zhang, Motilal, Bailey and Meinhardt2016). Around 24,000 cacao accessions, including wild and improved materials, are conserved in two international gene banks in Trinidad and Costa Rica and national research institutes such as CEPEC in Brazil (Lopes et al., Reference Lopes, Reis Monteiro, Pires, Clement, Yamada and Gramacho2011; Kodoth, Reference Kodoth2021). In Colombia, the collection conserved in the Corporación Colombiana de Investigación Agropecuaria (Agrosavia) includes around 600 wild and improved accessions (Rodriguez-Medina et al., Reference Rodriguez-Medina, Caicedo Arana, Sounigo, Argout, Alvarado and Yockteng2019). Despite the extensive collections and high genetic diversity conserved, most breeding programmes used a narrow genetic base to improve yield and resistance to pests and diseases (DuVal et al., Reference DuVal, Gezan, Mustiga, Stack, Marelli, Chaparro, Livingstone, Royaert and Motamayor2017; Rodriguez-Medina et al., Reference Rodriguez-Medina, Caicedo Arana, Sounigo, Argout, Alvarado and Yockteng2019; Ceccarelli et al., Reference Ceccarelli, Lastra, Loor Solórzano, Chacón, Nolasco, Sotomayor Cantos, Plaza Avellán, López, Fernández Anchundia, Dessauw, Orozco-Aguilar and Thomas2022; Daymond and Bekele, Reference Daymond, Bekele, Priyadarshan and Jain2022).
The study of crop wild relatives (CWR) is vital for agriculture and food security because they contain high levels of genetic diversity compared to cultivated crops and, as a result, can adapt to a wide range of habitats and environments, making them valuable for crop improvement (Maxted et al., Reference Maxted, Ford-Lloyd B, Jury, Kell and Scholten2006, Reference Maxted, Kell, Toledo, Dulloo, Heywood, Hodgkin, Hunter, Guarino, Jarvis and Ford-Lloyd2010, Reference Maxted, Kell, Ford-Lloyd, Dulloo and Toledo2012; Heywood et al., Reference Heywood, Casas, Ford-Lloyd, Kell and Maxted2007; Vincent et al., Reference Vincent, Wiersema, Kell, Fielder, Dobbie, Castañeda-Álvarez, Guarino, Eastwood, Leόn and Maxted2013; Zhang et al., Reference Zhang, Mittal, Leamy, Barazani and Song2017; Majeed et al., Reference Majeed, Chaudhary, Hulse-Kemp, Azhar and Azhar2021). Therefore, plant breeders should use a broader range of genetic resources, particularly CWR, that can provide new sources of agronomic traits to develop materials that respond to present and future challenges of the crop, including climate resilience. Collecting, conserving, characterizing and using CWR genetic resources are crucial to support breeding efforts (Maxted et al., Reference Maxted, Kell, Toledo, Dulloo, Heywood, Hodgkin, Hunter, Guarino, Jarvis and Ford-Lloyd2010; Ford-Lloyd et al., Reference Ford-Lloyd B, Schmidt, Armstrong, Barazani, Engels, Hadas, Hammer, Kell, Kang, Khoshbakht, Li, Long, Lu, Ma, Nguyen, Qiu, Ge, Wei, Zhang and Maxted2011; Dempewolf et al., Reference Dempewolf, Baute, Anderson, Kilian, Smith and Guarino2017; Ceccarelli et al., Reference Ceccarelli, Lastra, Loor Solórzano, Chacón, Nolasco, Sotomayor Cantos, Plaza Avellán, López, Fernández Anchundia, Dessauw, Orozco-Aguilar and Thomas2022; Renzi et al., Reference Renzi, Coyne, Berger, von Wettberg, Nelson, Ureta, Hernández, Smýkal and Brus2022). An effective conservation of cacao plant genetic resources in developing countries will need international and regional efforts. For instance, the Global Cacao Genetic Resources Network (CacaoNet) coordinated by Bioversity International aims to optimize the conservation and use of cacao genetic resources worldwide for the benefit of breeders, researchers and farmers (CacaoNet, Reference Laliberté2012).
Understanding the genetic diversity, population structure and genetic pedigree of cacao collections is crucial for conservation and breeding strategies. The molecular markers, simple-sequence repeats (SSR), have been extensively used to screen cacao germplasm (Borrone et al., Reference Borrone, Brown, Kuhn, Motamayor and Schnell2007; Zhang et al., Reference Zhang, Mischke, Johnson, Phillips-Mora and Meinhardt2009; Aikpokpodion et al., Reference Aikpokpodion, Kolesnikova-Allen, Adetimirin, Guiltinan, Eskes, Motamayor and Schnell2010; Irish et al., Reference Irish, Goenaga, Zhang, Schnell, Brown and Motamayor2010) since they have been reported as an international standard for cacao DNA fingerprinting (Lanaud et al., Reference Lanaud, Risterucci, Pieretti, Falque, Bouet and Lagoda1999; Pugh et al., Reference Pugh, Fouet, Risterucci, Brottier, Abouladze, Deletrez, Courtois, Clement, Larmande, N'Goran and Lanaud2004; Saunders et al., Reference Saunders, Mischke, Leamy and Hemeida2004). In 2008, Motamayor et al. (Reference Motamayor, Lachenaud, Wallace, Loor, Kuhn, Brown, Schnell, da Silva e Mota, Loor, Kuhn, Brown and Schnell2008) showed that the diversity of cacao based on 96 SSRs could be classified into 10 different genetic groups using collections from the Upper Amazon, Lower Amazon, Orinoco, north of South America, Central America and Guyana. In addition, Zhang et al. (Reference Zhang, Martínez, Johnson, Somarriba, Phillips-Mora, Astorga, Mischke and Meinhardt2012), using 15 SSRs, reported a new genetic group from Bolivia with a unique genetic profile different from the groups reported by Motamayor et al. (Reference Motamayor, Lachenaud, Wallace, Loor, Kuhn, Brown, Schnell, da Silva e Mota, Loor, Kuhn, Brown and Schnell2008). However, SSR markers have disadvantages, such as expensive and time-consuming processes and data sharing complications due to platform-to-platform variation (Livingstone et al., Reference Livingstone, Motamayor, Schnell, Cariaga, Freeman, Meerow, Brown and Kuhn2011).
Single nucleotide polymorphism (SNP) is the most common form of DNA sequence variation between alleles and is considered an ideal codominant marker system for assessing genetic diversity in model and non-model species (Batley and Edwards, Reference Batley and Edwards2007; Mammadov et al., Reference Mammadov, Aggarwal, Buyyarapu and Kumpatla2012). SNP markers are advantageous because they are abundant, ubiquitous, amenable to high- and ultra-high-throughput automation and have a low error rate compared with SS (Mammadov et al., Reference Mammadov, Aggarwal, Buyyarapu and Kumpatla2012). To study the identity of cacao, Livingstone et al. (Reference Livingstone, Royaert, Stack, Mockaitis, May, Farmer, Saski, Schnell, Kuhn and Motamayor2015) identified 330,000 SNPs using RNA-seq data from 16 diverse cacao cultivars and generated a 6 K SNP array. Other applications of SNP markers in cacao include genetic diversity analysis (Ji et al., Reference Ji, Zhang, Motilal, Boccara, Lachenaud and Meinhardt2013; Fang et al., Reference Fang, Meinhardt, Mischke, Bellato, Motilal and Zhang2014; Cosme et al., Reference Cosme, Cuevas, Zhang, Oleksyk and Irish2016; Osorio-Guarín et al., Reference Osorio-Guarín, Berdugo-Cely, Coronado, Zapata, Quintero, Gallego-Sánchez and Yockteng2017, Reference Osorio-Guarín, Quackenbush and Cornejo2018; Gopaulchan et al., Reference Gopaulchan, Motilal, Bekele, Clause, Ariko, Ejang and Umaharan2019, Reference Gopaulchan, Motilal, Kalloo, Mahabir, Moses, Joseph and Umaharan2020; Mahabir et al., Reference Mahabir, Motilal, Gopaulchan, Ramkissoon, Sankar and Umaharan2020; Wang et al., Reference Wang, Motilal, Meinhardt, Yin and Zhang2020), marker–trait association studies (Romero Navarro et al., Reference Romero Navarro, Phillips-Mora, Arciniegas-Leal, Mata-Quirós, Haiminen, Mustiga, Livingstone, van Bakel, Kuhn, Parida, Kasarskis, Motamayor, Romero-Navarro, Phillips-Mora, Arciniegas-Leal, Mata-Quirós, Haiminen, Mustiga, Livingstone, van Bakel, Kuhn, Parida, Kasarskis and Motamayor2017; McElroy et al., Reference McElroy, Navarro, Mustiga, Stack, Gezan, Peña, Sarabia, Saquicela, Sotomayor, Douglas, Migicovsky, Amores, Tarqui, Myles and Motamayor2018; Osorio-Guarín et al., Reference Osorio-Guarín, Berdugo-Cely, Coronado-Silva, Baez, Jaimes and Yockteng2020; Gutiérrez et al., Reference Gutiérrez, Puig, Phillips-Mora, Bailey, Ali, Mockaitis, Schnell, Livingstone, Mustiga, Royaert and Motamayor2021) and domestication studies (Cornejo et al., Reference Cornejo, Yee, Dominguez, Andrews, Sockell, Strandberg, Livingstone, Stack, Romero, Umaharan, Royaert, Tawari, Ng, Gutierrez, Phillips, Mockaitis, Bustamante and Motamayor2018).
Central America was considered the first centre of cacao domestication in Mesoamerica about 1900 years ago (Miranda, Reference Miranda1962). However, a recent study showed that cacao was cultivated earlier (5300 years ago) in the north-western part of the Amazonia region, predominantly in southern Ecuador (Zarrillo et al., Reference Zarrillo, Gaikwad, Lanaud, Powis, Viot, Lesur, Fouet, Argout, Guichoux, Salin, Solorzano, Bouchez, Vignes, Severts, Hurtado, Yepez, Grivetti, Blake and Valdez2018). In Colombia, the expansion of cacao cultivation occurred in the 17th century in the northeastern region (Guerrero-Rincón et al., Reference Guerrero-Rincón, Pabón and Ferreira1998). Besides, efforts in the country to conserve cacao diversity have been made since the 1940s to safeguard farmers' livelihoods and preserve food security with genetic material introduced from other countries and some combinations with native cacao (Rodriguez-Medina et al., Reference Rodriguez-Medina, Caicedo Arana, Sounigo, Argout, Alvarado and Yockteng2019). Colombia has high diversity of cacao CWR (González-Orozco et al., Reference González-Orozco, Galán, Ramos and Yockteng2020) and high genetic diversity of cultivated cacao based on random amplified microsatellites (RAM) markers (H e: 0.28) regionally (Morillo et al., Reference Morillo, Morillo, Muñoz, Ballesteros and González2014) and countrywide (H e: 0.314) based on SNP markers (Osorio-Guarín et al., Reference Osorio-Guarín, Berdugo-Cely, Coronado, Zapata, Quintero, Gallego-Sánchez and Yockteng2017). High phenotypic variability using morpho-agronomic descriptors related to productivity, flower and seed traits has also been reported (Ballesteros et al., Reference Ballesteros, Lagos and L2016; López-Hernández et al., Reference López-Hernández, Sandoval-Aldana, García-Lozano and Criollo-Nuñez2021).
In addition to the genetic diversity and phenotypic studies, it is essential to investigate the phylogenetic relationships of current genotypes. The reconstruction of a phylogenetic tree can untangle the relationship between genotypes, and phylogenetic diversity (PD) can improve our understanding of evolutionary events that determine the current diversity within a species (Faith and Baker, Reference Faith and Baker2006; Kapli et al., Reference Kapli, Yang and Telford2020). The most traditional biodiversity metric, species richness, only considers the number of species. In contrast, PD provides a comparable, evolutionary measure of biodiversity not possible with species counts (Miller et al., Reference Miller, Jolley-Rogers, Mishler and Thornhill2018). PD is the sum of the branch lengths of a tree that connects all studied species (Faith, Reference Faith1992). This measure can be applied to any taxon regardless of its origin or rank (Fisher et al., Reference Fisher, Wall, Yip and Mishler2007; Mishler, Reference Mishler2021) and is widely used in plant conservation, crop science, biogeography, biodiversity and climate change (González-Orozco et al., Reference González-Orozco, Mishler, Miller, Laffan, Knerr, Unmack, Georges, Thornhill, Rosauer and Gruber2015; Laity et al., Reference Laity, Laffan, González-Orozco, Faith, Rosauer, Byrne, Miller, Crayn, Costion, Moritz and Newport2015; Nagalingum et al., Reference Nagalingum, Knerr, Laffan, González-Orozco, Thornhill, Miller and Mishler2015; Laffan et al., Reference Laffan, Rosauer, Di Virgilio, Miller, González-Orozco, Knerr, Thornhill and Mishler2016; Thornhill et al., Reference Thornhill, Mishler, Knerr, González-Orozco, Costion, Crayn, Laffan and Miller2016). Using PD as a criterion in conservation planning could reduce the risk of losing entire groups or lineages (Soulebeau et al., Reference Soulebeau, Pellens, Lowry, Aubriot, Evans, Haevermans, Pellens and Grandcolas2016).
Colombia is a potential source of unexplored cacao CWR diversity (González-Orozco et al., Reference González-Orozco, Sosa, Thornhill and Laffan2021), making it a research priority for the in situ conservation of native genetic resources. Areas of high PD could be a potential source of genetic resources well-adapted and resilient to modern challenges due to the climate change that can be used in breeding programmes (González-Orozco et al., Reference González-Orozco, Sosa, Thornhill and Laffan2021).
Osorio-Guarín et al. (Reference Osorio-Guarín, Berdugo-Cely, Coronado, Zapata, Quintero, Gallego-Sánchez and Yockteng2017) explored the phylogenetic relationships among cacao genotypes using SNP markers. However, the authors used this analysis to check if the Colombian germplasm collection represents the diversity of the species. To the best of our knowledge, this study is the first to apply PD to understand better the diversity of Colombian cacao in Agrosavia national germplasm dataset. Our study is pioneering because it combines information on its geographical distribution and evolutionary relationships to contribute to selecting in situ priority areas and ex situ management strategies of cacao germplasm in Colombia.
Materials and methods
Plant material
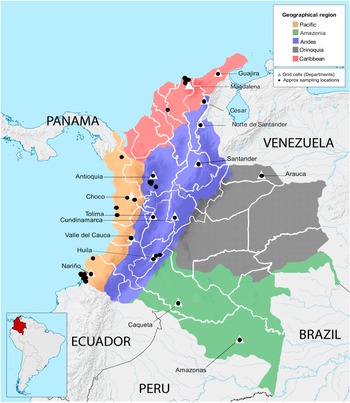
A total of 279 wild and cultivated accessions conserved in the Corporación Colombiana de Investigación Agropecuaria (Agrosavia) germplasm collection were evaluated (online Supplementary Table S1). These accessions are stored ex situ at the Agrosavia research centre in Palmira (3°30′41″N 76°19′19″W). The wild accessions (179) were collected from habitats in the following departments: Magdalena, Guajira, Cesar, Norte de Santander, Nariño, Choco and Amazonas. The cultivated accessions (100) came from agricultural areas in the departments of Arauca, Valle del Cauca, Huila, Tolima, Cundinamarca, Antioquia and Santander (Fig. 1, online Supplementary Table S1).
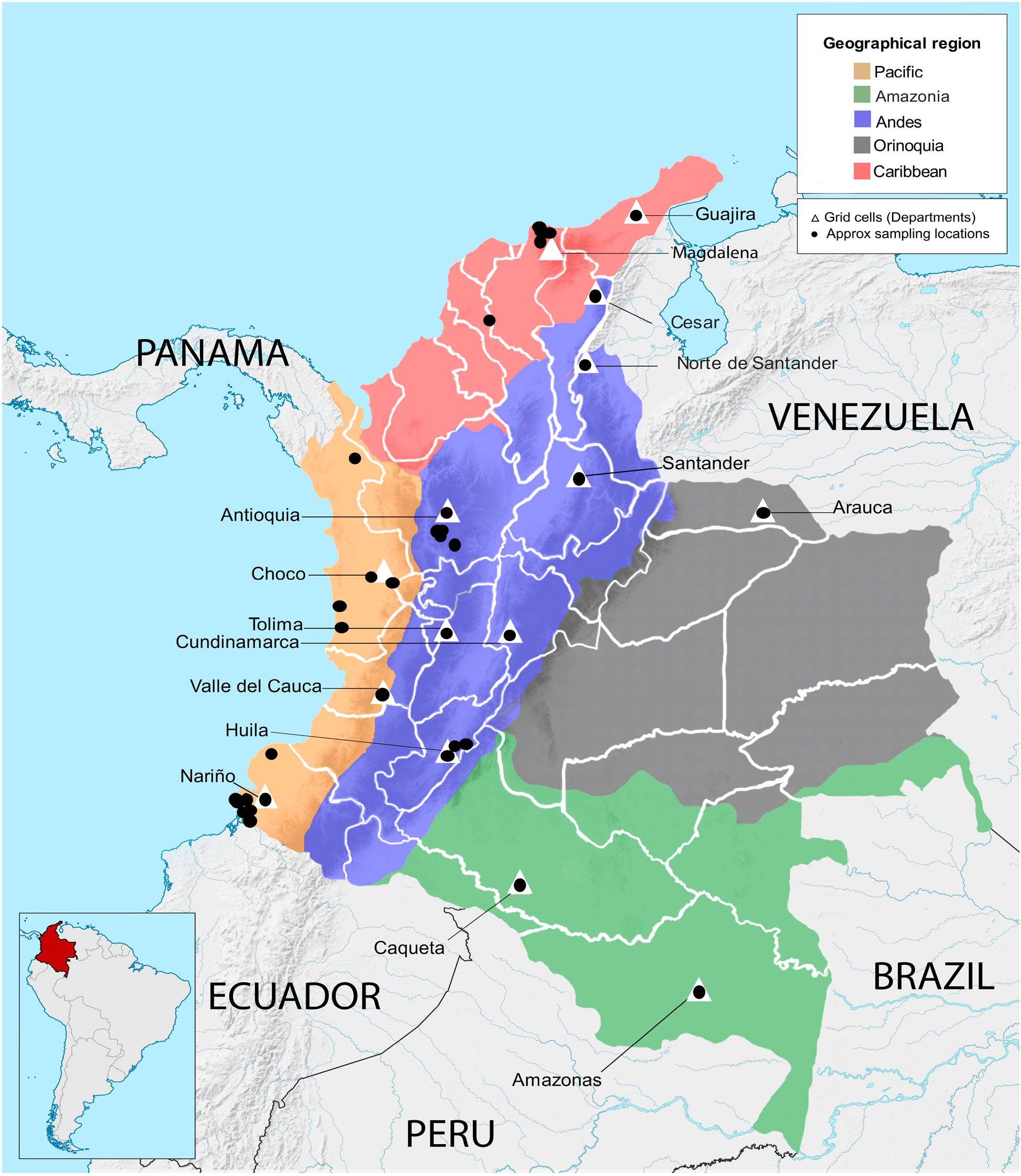
Fig. 1. Distribution of collection sites of Theobroma cacao genotypes across geographical regions of Colombia. Names of Colombian departments in which samples were collected are indicated. Triangles indicate grid cells used for the phylogenetic diversity (PD) analysis.
Phylogenetic and genetic diversity analyses
We used an alignment of 87 SNPs (Osorio-Guarín et al., Reference Osorio-Guarín, Berdugo-Cely, Coronado, Zapata, Quintero, Gallego-Sánchez and Yockteng2017) and their flanking invariant region of 60 bp to avoid branch length bias in the phylogenetic analysis. This subset of SNPs belongs to an original set composed of 1560 candidate SNPs developed from cDNA sequences in cacao tissues, more specifically, flowers, cherelles, pod cortex, shoots, roots, germinated seeds and embryos from an in vitro culture (Argout et al., Reference Argout, Fouet, Wincker, Gramacho, Legavre, Sabau, Risterucci, Da Silva, Cascardo, Allegre, Kuhn, Verica, Courtois, Loor, Babin, Sounigo, Ducamp, Guiltinan, Ruiz, Alemanno, Machado, Phillips, Schnell, Gilmour, Rosenquist, Butler, Maximova and Lanaud2008; Allegre et al., Reference Allegre, Argout, Boccara, Fouet, Roguet, Bérard, Thévenin, Chauveau, Rivallan, Clement, Courtois, Gramacho, Boland-Augé, Tahi, Umaharan, Brunel and Lanaud2012). Panel selection was based on an SNP call rate percentage higher than 90%, represented across 10 cacao chromosomes and heterozygosity results (Ji et al., Reference Ji, Zhang, Motilal, Boccara, Lachenaud and Meinhardt2013; Fang et al., Reference Fang, Meinhardt, Mischke, Bellato, Motilal and Zhang2014; Osorio-Guarín et al., Reference Osorio-Guarín, Berdugo-Cely, Coronado, Zapata, Quintero, Gallego-Sánchez and Yockteng2017). The protocol for SNP genotyping of cacao uses the Fluidigm 96.96 Dynamic ArrayTM (Fluidigm, San Francisco, CA, USA) (Osorio-Guarín et al., Reference Osorio-Guarín, Berdugo-Cely, Coronado, Zapata, Quintero, Gallego-Sánchez and Yockteng2017).
The nucleotide sequences of all targeting SNPs were aligned in ClustalX v1.83 (Larkin et al., Reference Larkin, Blackshields, Brown, Chenna, McGettigan, McWilliam, Valentin, Wallace, Wilm, Lopez, Thompson, Gibson and Higgins2007). After concatenating the data, an evolutionary model was estimated using the option SMS of PhyML software (Lefort et al., Reference Lefort, Longueville and Gascuel2017). The phylogenetic tree was constructed by computing 1000 bootstrap replicates using the maximum likelihood (ML) method in the PhyML v3.0 program (Guindon et al., Reference Guindon, Dufayard, Lefort, Anisimova, Hordijk and Gascuel2010) found in a bioinformatics platform (http://www.atgc-montpellier.fr/phyml/). Gap sites were treated as missing data. We also conducted a Bayesian analysis using MrBayes v3.2 (Huelsenbeck and Ronquist, Reference Huelsenbeck and Ronquist2001) in CIPRES (Miller et al., Reference Miller, Pfeiffer and Schwartz2010). Using the metropolis-coupled Markov chain Monte Carlo (MCMC) algorithm, two independent runs of 50 million generations were sampled for one in every 1000 trees. The results of the MrBayes analysis were examined for convergence of parameters using Tracer v1.7 (Rambaut et al., Reference Rambaut, Suchard, Xie and Drummond2018), excluding the initial 10% of MCMCs. Posterior probabilities of clades were obtained from the 50% majority rule consensus of the sampled trees. The accession copoazu_75 from the species Theobroma grandiflorum (Wild Ex. Spreng. Schum), commonly known as copoazu, cupuassu or cacao blanco, was used as an outgroup. The ancestral distribution of Colombian cacao genotypes was reconstructed using the parsimony method with the software Mesquite v3.61 (Maddison and Maddison, Reference Maddison and Maddison2021).
Finally, SNPs were scored as codominant markers with the software GenAlex v6.5 (Peakall and Smouse, Reference Peakall and Smouse2006, Reference Peakall and Smouse2012) to perform two further analyses: (1) standard measures of genetic diversity such as the effective number of alleles per locus (N e), expected heterozygosity (H e) and observed heterozygosity (H o) for each department; and (2) genetic differentiation via covariance matrix with data standardization among populations based on G-Statistics (Jost's D EST) (Jost, Reference Jost2008) visualized through a principal coordinate analysis (PCoA).
Phylogenetic diversity analyses
The spatial analysis tool in the software Biodiverse v1.0 (http://shawnlaffan.github.io/biodiverse/) (Laffan et al., Reference Laffan, Lubarsky and Rosauer2010) was used to calculate genotype richness (GR), PD, and relative phylogenetic diversity (RPD) at a spatial resolution of 1 degree, corresponding to 100 × 100 km. We used 279 geolocations of cacao genotypes from different geographical regions of Colombia (online Supplementary Table S2). GR is the number of genotypes in a grid cell referred to as observed GR (Laffan et al., Reference Laffan, Rosauer, Di Virgilio, Miller, González-Orozco, Knerr, Thornhill and Mishler2016), and PD is an observed measure of diversity, calculated as the total sum of branch lengths (in this case, each branch represents a cacao genotype) in each cell (Faith, Reference Faith1992). RPD is a measure of PD using a standardization and randomization process to avoid bias caused by the number of taxa in a cell (Mishler et al., Reference Mishler, Knerr, González-Orozco, Thornhill, Laffan and Miller2014). RPD was calculated using the ratio of observed PD and a comparison tree with the same topology and equal branch lengths (Mishler et al., Reference Mishler, Knerr, González-Orozco, Thornhill, Laffan and Miller2014).
In addition, observed PD values were compared to the expected values using a randomization test of 999 iterations completed in the ¨rand_structured¨ model in Biodiverse v1.0. A two-tailed test with an α of 0.05 was applied to obtain the significance of the observed values compared to the expected values. The PD randomization test produced the significance test for the observed PD, referred to as significant phylogenetic diversity (SPD). The Colombian map, which displayed the significant values, was generated using R scripts (R Core Team and R development core team, 2008). Codes are available at https://github.com/NunzioKnerr/biodiverse_pipeline.
Results
Phylogenetic and genetic diversity analyses
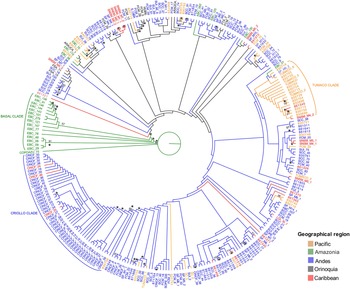
The concatenation of the SNPs with their flanking regions produced an alignment with a total length of 21,054 bp. The Bayesian and likelihood analyses produced a similar topology using the evolution general time reversible + gamma + invariable sites (GTR + G + I), model, with a proportion of invariable sites of 0.968 and a γ shape parameter of 0.062. The reconstruction of the ancestral distribution of Colombian cacao genotypes showed that the Amazonia region was an ancestral centre of distribution (Fig. 2). A group of botanical expedition of caqueta (EBC) accessions collected in the Amazonia region were found to be the earliest diverging lineages in the tree. Some Pacific accessions collected near Tumaco (Nariño) formed a small clade (Fig. 2). Genotypes from the Andes and Caribbean regions were distributed across the phylogenetic tree. The exception was the criollo corpoica fedecacao (CRICF) accessions collected in Cesar from the Andes region, regrouping into a separate supported clade in a derived position (Fig. 2).

Fig. 2. Phylogeny of Theobroma cacao genotypes of Colombia with reconstruction of geographical distribution of genotypes. Bootstrap values higher than 50% are indicated in nodes and posterior probabilities higher than 0.9 are indicated by an asterisk (*). The basal clade corresponds to EBC accessions collected in the Amazonia region, the Tumaco clade corresponds to accessions collected from this location in the Nariño department in the Pacific region and Criollo clade corresponds to CRICF accessions collected in the Cesar department in the Andes region.
To avoid bias, we calculated the genetic diversity indices H e and H o, excluding departments with just one sample (Caquetá, Cundinamarca, Guajira and Tolima). The genetic analysis showed that H o ranged from 0.151 to 0.467 with a mean value of 0.358, while H e ranged from 0.242 to 0.422 with an average value of 0.367 (Table 1). The results showed that H o is lower than H e for Amazonas, Cesar, Chocó, Huila and Magdalena. In contrast, Antioquia, Arauca, Nariño, Norte de Santander, Santander and Valle del Cauca showed higher H o than H e.
Table 1. Summary of the statistics of genetic diversity calculated with 87 SNP for 275 Theobroma cacao accessions

The PCoA was applied to visualize and investigate the differentiation among populations. It was necessary to remove accessions from Arauca, Caquetá, Cundinamarca, Choco, Guajira, Huila, Tolima and Valle del Cauca because the D EST analysis needs at least 20 individuals per population (Gerlach et al., Reference Gerlach, Jueterbock, Kraemer, Deppermann and Harmand2010). The first principal coordinate accounted for 84.73%, while the second explained 6.23% of the variation, together accounting for 90.96% of the total variation (Fig. 3). The PCoA distinguished Norte de Santander (Serranía de los Motilones), Antioquia and Magdalena (Sierra Nevada de Santa Marta) as the most differentiated locations, while Amazonas, Santander, Cesar and Nariño clustered together on the left side of the biplot. The differentiation was significant (P < 0.05) and ranged between 0.001 and 0.555 using 999 permutations.

Fig. 3. Principal coordinate analysis (PCoA) run on GenAlEx using the DEST pairwise distances.
Phylogenetic diversity analyses
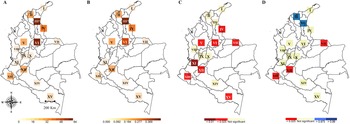
The 279 geolocations of cacao genotypes were mapped in 15 PD grid cells (triangles in Fig. 1). As anticipated, GR was significantly related to PD (r 2: 0.7955; online Supplementary Fig. S1). The highest observed PD (about 46%) and GR (68 of the 279 genotypes) were found in the Serranía del Perijá, Cesar, northern Colombia (Fig. 4(a), III*). Areas of high observed PD were also found near the northern tip of the eastern Andes range, the geographically isolated mountains of the Sierra Nevada de Santa Marta, Serranía del Perijá and Serranía de Los Motilones (Fig. 4(b)).

Fig. 4. Observed and randomised diversity patterns of Theobroma cacao genotypes in Colombia. (a) Genotypes richness (GR), (b) phylogenetic diversity (PD), (c) significant phylogenetic diversity (SPD), (d) relative phylogenetic diversity (RPD). The sites of interest are numbered as follows: I: Guajira; II: Sierra Nevada de Santa Marta in Magdalena department; III: Serranía del Perijá in Cesar department; IV: Serranía de los Motilones in Norte de Santander department; V: Antioquia; VI: Santander; VII: Arauca; VIII: Choco; IX: Tolima; X: Cundinamarca; XI: Valle del Cauca; XII: Huila; XIII: Nariño; XIV: Caquetá and XV: Amazonas. The asterisk indicates grid cells with the highest observed values of GR and PD, respectively.
The second-highest concentration of PD and GR was found in the cacao-producing region in Santander, with a PD of 35.2% (Fig. 4(a) and (b), VI). PD was significant after a randomization test (SPD), demonstrating that the diversity found is more closely related than expected by chance (Fig. 4(c)). Using this analysis, Norte de Santander, Antioquia, Santander, Arauca, Valle del Cauca, Huila, Nariño and Amazonas show a significant SPD, indicating that these regions are diverse (Fig. 4(c)).
Areas of significantly high RPD included the Sierra Nevada de Santa Marta and the Serranía del Perijá in northeast Colombia (Fig. 4(d)). On the contrary, Arauca, Huila and Nariño had a significantly low RPD (Fig. 4(d)).
Discussion
A previous study of Colombian cacao genotypes (Osorio-Guarín et al., Reference Osorio-Guarín, Berdugo-Cely, Coronado, Zapata, Quintero, Gallego-Sánchez and Yockteng2017) reported high genetic diversity in the germplasm collection but did not consider the PD between materials. The present study is the first to locate centres of PD of Colombian cacao genotypes and disentangle their evolutionary relationships. Our aim was not to reconstruct historical evidence of evolutionary processes but to find a reliable way to validate the diversity of Colombian cacao based on spatial distribution patterns and form a basis for guiding further sampling and increasing the cacao gene pool available for breeding and crop improvement.
The results showed that H o is lower than H e for Amazonas, Cesar, Chocó, Huila and Magdalena, indicating excess homozygosity explained by inbreeding or isolation by distance in this cacao population. In contrast, Antioquia, Arauca, Nariño, Norte de Santander, Santander and Valle del Cauca showed higher H o than H e, resulting in an excess of heterozygosity, most likely because these locations are cacao-producing regions in which genotypes have been crossbred indiscriminately. We found that the most diverse regions based on H e values were Magdalena, Nariño, Santander and Norte de Santander. These results can be due to the fact that Santander and Norte de Santander are Colombia's most important producing regions where breeding programmes have been carried out, while Nariño and Magdalena are considered to have regional materials of hybrid origin (Ballesteros et al., Reference Ballesteros, Lagos and L2016; Ramos Ospino et al., Reference Ramos Ospino, Gómez Alvaréz, Machado-Sierra and Aranguren2020).
The phylogenetic analysis showed low bootstrap values; however, the analysis was performed using two reconstruction methods (ML and Bayesian), resulting in similar topologies and giving robustness to our interpretations. Colombian cacao genotypes result from many historical events, including hybridization (Rodriguez-Medina et al., Reference Rodriguez-Medina, Caicedo Arana, Sounigo, Argout, Alvarado and Yockteng2019) that may cause low support of nodes, reported as a problem in phylogenetic studies (McDade, Reference McDade1990). In the case of Criollo group, the low bootstrap support would be due to the high level of homozygosity (Motamayor et al., Reference Motamayor, Risterucci, Lopez, Ortiz, Moreno and Lanaud2002). We found a group of Amazonia genotypes (EBC) positioned at the root of the phylogenetic tree (Fig. 2), which is indicative of the ancestral origin and agrees with the studies that recognize the Amazonia region as the centre of origin of cultivated and wild species (Thomas et al., Reference Thomas, van Zonneveld, Loo, Hodgkin, Galluzzi and van Etten2012; Zarrillo et al., Reference Zarrillo, Gaikwad, Lanaud, Powis, Viot, Lesur, Fouet, Argout, Guichoux, Salin, Solorzano, Bouchez, Vignes, Severts, Hurtado, Yepez, Grivetti, Blake and Valdez2018). The EBC genotypes were collected from the Amazonia region during an expedition to the low parts of the Caquetá river (Allen, Reference Allen1988). Recently, a study including all the cacao genetic groups showed that samples of EBC genotypes are related to the Ecuadorian group Curaray at the base of the phylogeny (Osorio-Guarín et al., Reference Osorio-Guarín, Berdugo-Cely, Coronado, Zapata, Quintero, Gallego-Sánchez and Yockteng2017). In addition, a genetic structure analysis based on 9000 SNPs showed that the EBC-06, 09, 29 and EBC-48 have more than 90% of Curaray ancestry (Osorio-Guarín et al., Reference Osorio-Guarín, Berdugo-Cely, Coronado-Silva, Baez, Jaimes and Yockteng2020).
We also recovered a clade that regroups most of the Criollo (CRICF) genotypes, a genetic group previously reported as a differentiated one by Motamayor et al. (Reference Motamayor, Lachenaud, Wallace, Loor, Kuhn, Brown, Schnell, da Silva e Mota, Loor, Kuhn, Brown and Schnell2008). Half of the regional genotypes from Tumaco (Nariño) formed a clade in our phylogenetic tree, probably indicating the distinctiveness of some of the Tumaco materials. This region produces high-quality cacao, winning international prizes, including the Cocoa for Excellence in 2015 (Montoya-Restrepo et al., Reference Montoya-Restrepo, Montoya-Restrepo and Lowy-Ceron2015; Arango, Reference Arango2017). Most genotypes from Nariño are phylogenetically related because they formed tighter groups of closely related branches, a pattern known as phylogenetic clustering (Webb et al., Reference Webb, Ackerly, McPeek and Donoghue2002). The genetic structure analysis of these samples showed that most of their ancestry is related to a mix of the Nacional, Criollo and Amelonado genetic groups (Osorio-Guarín et al., Reference Osorio-Guarín, Berdugo-Cely, Coronado-Silva, Baez, Jaimes and Yockteng2020).
Genotypes from the Sierra Nevada de Santa Marta (Magdalena), Serranía de Los Motilones (Norte de Santander) and Serranía del Perijá (Cesar) (excluding some of the CRICF genotypes) were distributed across the tree, suggesting that the genotypes from these sites are distantly related and do not share a close common ancestor. Patiño Rodríguez (Reference Patiño Rodríguez2002) mentioned that in the 1600s, the most crucial region for cacao cultivation was northeastern Colombia (Norte de Santander). Later in the 1950s, the south Pacific region (Valle del Cauca) was the central cacao-producing region, explaining the presence of non-phylogenetic-related cacao genotypes (Patiño Rodríguez, Reference Patiño Rodríguez2002). In these two cases, we observed long and distantly related branches, a pattern known as phylogenetic overdispersion (Webb et al., Reference Webb, Ackerly, McPeek and Donoghue2002). Cacao materials with desired agronomic traits from different sources were probably transported to these producing regions, explaining why they are not necessarily closely related. The rest of the genotypes mostly come from the Andes region and are distributed in different shallow clades across the tree (Fig. 2), showing a mixed pattern. For example, some genotypes from Norte de Santander and Antioquia are strongly related (phylogenetic clustering), and some are broadly distributed across the tree (phylogenetic overdispersion).
Areas with a high GR often coincide with areas with high PD, as we found in our study (Fig. 4(a) and (b)). Other studies have also reported a significant correlation (Mishler et al., Reference Mishler, Knerr, González-Orozco, Thornhill, Laffan and Miller2014; Qian et al., Reference Qian, Deng, Jin, Mao, Zhao and Ricklefs2019; Manish, Reference Manish2021). However, an index such as GR cannot explain the complexity of the evolutionary events causing the current diversity of taxa. For instance, a study of the flora biodiversity hotspots of the Cape Peninsula in Africa that explored the utility of these indices showed that it is more beneficial to use a decoupling PD from GR because this complex diversity has a solid phylogeographic structure as a consequence of endemic radiations (Forest et al., Reference Forest, Grenyer, Rouget, Davies, Cowling, Faith, Balmford, Manning, Procheş, van der Bank, Reeves, Hedderson and Savolainen2007). For our study, we applied various indices to ensure diversity was understood in different forms.
Observed biodiversity patterns can be deceptive because of different sampling biases (Swenson, Reference Swenson2009; Schmidt-Lebuhn et al., Reference Schmidt-Lebuhn, Knerr and González-Orozco2012; Tucker and Cadotte, Reference Tucker and Cadotte2013), such as the effect of remoteness on the sampling of field collections. For instance, genotypes from the Pacific, Orinoquia and Amazonia regions, which are remote and difficult to access, are under-represented. Updated biodiversity indices such as randomized RPD have been developed to examine the over-representation of long and deep branches and avoid this bias, which produces significantly high RPD values and is related to phylogenetic overdispersion. The over-representation of short or shallow branches produces significantly low RPD values related to phylogenetic clustering in the tree (Mishler et al., Reference Mishler, Knerr, González-Orozco, Thornhill, Laffan and Miller2014; Laffan et al., Reference Laffan, Rosauer, Di Virgilio, Miller, González-Orozco, Knerr, Thornhill and Mishler2016).
We found that the Sierra Nevada de Santa Marta in Magdalena and the Serranía del Perijá in Cesar exhibited significantly high RPD, possibly due to the geographic isolation of these cacao populations (blue grid cells in Fig. 4(d)). Significantly high RPD (phylogenetic overdispersion) can be explained by the occurrence of genotypes in an area containing relicts from past climate change or by a strong environmental heterogeneity that makes different niches available (Mayfield and Levine, Reference Mayfield and Levine2010; de Bello et al., Reference de Bello, Vandewalle, Reitalu, Lepš, Prentice, Lavorel and Sykes2013). It can also be explained by competition between CWRs that do not permit their co-occurrence in the same place (Webb et al., Reference Webb, Ackerly, McPeek and Donoghue2002).
The Sierra Nevada de Santa Marta (Magdalena), the Serranía del Perijá (Cesar), and the Serranía de los Motilones (Norte de Santander) are mountains disconnected from the Andes Mountain range. These regions contain long branches, implying the conglomeration of genotypes that possibly diversified recently with some genotypes with an older history. Isolated mountain ranges create micro-niches with singular ecological and climate conditions (dry and hot) that provide unique environments for agriculture and a concentration of endemic species (Webb and Peart, Reference Webb and Peart2000; Webb et al., Reference Webb, Ackerly, McPeek and Donoghue2002; Cooper et al., Reference Cooper, Freckleton and Jetz2011). Our results agree with Bryant et al. (Reference Bryant, Lamanna, Morlon, Kerkhoff, Enquist and Green2008), who found that angiosperms are more phylogenetically dispersed at higher elevations. In concordance with these findings, the Jost D EST analysis showed that genotypes from the mountain ranges such as Serranía de Los Motilones (Norte de Santander) and Sierra Nevada de Santa Marta (Magdalena) are more differentiated.
The Serranía del Perijá is a hotspot recognized for its high plant endemism levels (Cuatrecasas, Reference Cuatrecasas1964; Rivera Díaz and Fernández Alonso, Reference Rivera Díaz and Fernández Alonso2003). Most of the CRICF genotypes in this site belong to the Criollo genetic group (Osorio-Guarín et al., Reference Osorio-Guarín, Berdugo-Cely, Coronado, Zapata, Quintero, Gallego-Sánchez and Yockteng2017), the most genetically differentiated cacao group not only morphologically but also in quality and taste (Motamayor et al., Reference Motamayor, Mockaitis, Schmutz, Haiminen, Livingstone, Cornejo, Findley, Zheng, Utro, Royaert, Saski, Jenkins, Podicheti, Zhao, Scheffler, Stack, Feltus, Mustiga, Amores, Phillips, Marelli, May, Shapiro, Ma, Bustamante, Schnell, Main, Gilbert, Parida and Kuhn2013). Despite its distinctiveness, the diversity (H e: 0.032 and polymorphic sites: 50%) of the Criollo group was found to be low in our study, agreeing with the results of Motamayor et al. (Reference Motamayor, Risterucci, Lopez, Ortiz, Moreno and Lanaud2002), which could explain the predominantly short branches in the phylogenetic tree, implying that a population bottleneck probably occurred in this region. The selection and inbreeding of a few individuals caused the reduced genetic diversity and the closely related genotypes.
In contrast, Arauca, Huila and Nariño had significantly low RPDs (phylogenetic clustering), likely due to the conglomeration of cacao lineages that have recently diverged and probably result from hybridization events because these sites are cacao-producing regions. Low RPD is explained by the recent divergence of lineages in an area or by the co-occurrence of close relatives in the same community, excluding weaker competitors (Mishler et al., Reference Mishler, Knerr, González-Orozco, Thornhill, Laffan and Miller2014). For example, Arauca genotypes are distributed in different clades with predominantly short branch lengths, suggesting a homogenization by excessive crosses of closely related genotypes. Most of the Arauca genotypes are selected regionally by the Federación Nacional de Cacaoteros (Fedecacao) based on agronomic traits. In the study of Osorio-Guarín et al. (Reference Osorio-Guarín, Berdugo-Cely, Coronado, Zapata, Quintero, Gallego-Sánchez and Yockteng2017), some Arauca materials have approximately 50% of Iquitos ancestry.
High PD, significantly low SPD and RPD values were found in the southern departments of Valle del Cauca and Huila (Fig. 4(b)), suggesting a more recent evolutionary history. Indigenous settlements in both regions have cultivated the lands for years (1–900 AD), which may explain this high diversity. The archaeological sites of San Agustin in Huila and Calima in Valle del Cauca are known as centres of diversity for different crops (Velandia Jagua, Reference Velandia Jagua, Politis and Alberti1999; Piperno et al., Reference Piperno, Ranere, Dickau and Aceituno2017). Early evidence of plant food production closely related to native wild ancestors of crops such as squash, arrowroot and cocoyam has also been found in the Calima Valley (Piperno, Reference Piperno2011). Recent evidence in the Amazonia region of Ecuador showed that cultivated cacao has its origins in ancient indigenous sites (Zarrillo et al., Reference Zarrillo, Gaikwad, Lanaud, Powis, Viot, Lesur, Fouet, Argout, Guichoux, Salin, Solorzano, Bouchez, Vignes, Severts, Hurtado, Yepez, Grivetti, Blake and Valdez2018). A similar situation could have happened in Colombia on indigenous sites under the assumption that they were centres of food exchange, including cacao.
Most of the cacao genotypes in Colombia are more closely related than expected by chance (Fig. 4(c) and (d)), which can indicate the degradation of the original wild diversity. These genotypes with short branches are probably the result of hybridization, which could be detrimental because of genetic homogenization (Olden et al., Reference Olden, Poff, Douglas, Douglas and Fausch2004; Tieman et al., Reference Tieman, Zhu, Resende, Lin, Nguyen, Bies, Rambla, Beltran, Taylor, Zhang, Ikeda, Liu, Fisher, Zemach, Monforte, Zamir, Granell, Kirst, Huang and Klee2017) and, therefore, endemic losses of gene pools (Charlesworth, Reference Charlesworth2003). Two hypotheses can explain genetic homogenization: (1) intense inbreeding of the same materials in modern manipulation of crops or (2) cacao cultivation in indigenous sites, causing interbreeding of older lineages.
Furthermore, the PD would generate baseline information to improve cultivated cacao, introducing new genetic resources in a breeding programme whose agronomic performance should be previously assessed. This germplasm would then broaden the gene pool and increase population variation to solve the problems of the crop.
Conclusions
The application of PD helps analyse genetic diversity in germplasm collections, understand the evolutionary relationships among cacao genotypes in Colombia and identify centres of diversity and conservation. It is necessary to prioritize areas climatically stable with over-dispersed genotypes and stressed environments with clustered genotypes to conserve Colombian cacao diversity. Unlike cacao genotypes found in most parts of Colombia with predominantly short and closely related branches, the Amazonia region features long and distantly related branches, making it a likely location for wild cacao diversity. Since samples from the Amazonia region are underrepresented, collection in this region should be a priority to increase relict diversity for further genetic improvements of cultivated cacao. As well as regions with a significantly low RPD, such as Arauca, Huila and Nariño, with a conglomeration of cacao, recently diverged lineages. The Caribbean and northern Andes regions are the main areas where PD and significantly high RPD tend to concentrate. Particularly, the North Andes regions of Magdalena, Cesar and Norte de Santander, which have both relict and recent cacao diversity, should be prioritized as conservation areas. Collecting germplasm from selected priority areas would improve ex-situ holdings, provide potential new diversity for cacao improvement and increase the genetic diversity in cultivated materials.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S1479262123000047
Acknowledgments
We thank Nunzio Knerr, Joe Miller, Mario Porcel and Gina Garzón for providing valuable comments on the manuscript. Special thanks to Allende Pesca and Orlando Guiza for sharing their insights on the history of cacao cultivation in Colombia. We thank Jhon Berdugo, Eliana Báez and Roberto Coronado for their involvement in developing the genotype dataset. The manuscript was proofread and edited by Julia Alice Veronica de Raadt.