Diet is an important modifiable factor that can affect DNA methylation, one of the most extensively studied epigenetic mechanisms. Studying the effects of diet on DNA methylation may provide insight into the underlying mechanisms by which diet induces changes in health status. There is a clear need to further develop the field of nutritional epigenetics in humans, yet current challenges and pitfalls need to be addressed.
In a recent publication by ElGendy et al.(Reference ElGendy, Malcomson and Lara1), the evidence for effects of dietary interventions on DNA methylation in adults was systematically reviewed. The authors provided a comprehensive overview of dietary randomised controlled trials including DNA methylation outcomes and evaluated the corresponding changes in DNA methylation. Although highly relevant and timely, this publication does demonstrate some pitfalls in the field that need to be pointed out.
One challenge of nutritional epigenetic studies is obtaining a sufficiently large sample size. As shown by the authors, the total sample size of the randomised controlled trials that were included varied from 7 to 388. Of the studies, 70 % included fewer than fifty participants (Table 1). Reassuringly, the small sample sizes do not seem to play a major role in creating false positives in the studies on this topic, as no significant trend was observed in changes for success according to sample size (Table 1). Nevertheless, future studies would benefit from larger sample sizes to increase the power to detect novel DNA methylation loci affected by diet.
Table 1. Number of studies included by ElGendy et al. (Reference ElGendy, Malcomson and Lara1) reporting a statistically significant effect, by sample size and DNA methylation approach
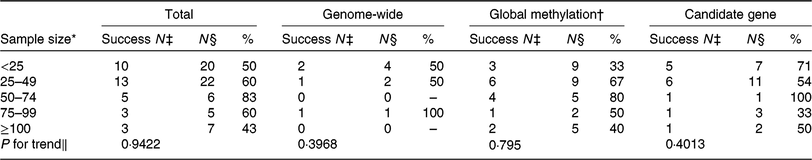
* Sample size in five categories for number of participants included.
† Although not labelled by ElGendy et al. ( Reference ElGendy, Malcomson and Lara1 ) as global methylation studies, LINE-1 pyrosequencing was included with the global methylation studies.
‡ Success N = number of studies reporting a statistically significant effect as per ElGendy et al.( Reference ElGendy, Malcomson and Lara1 ).
§ N = number of studies. A study is defined as an analysis of one DNA methylation approach.
∥ χ 2 P value for trend.
ElGendy et al. (Reference ElGendy, Malcomson and Lara1) stated that diverse DNA methylation assessment techniques have been used, which made it difficult to compare and combine data. Careful examination of the tables (ElGendy et al.(Reference ElGendy, Malcomson and Lara1), Tables 1–6) reveals that the diverse DNA methylation assessment techniques have led the authors to misclassify several of the included articles. Two studies labelled as ‘genome-wide’ should have been labelled ‘global [methylation]’, based on the DNA methylation technique used (methyl acceptance assay and liquid chromatography–mass spectrometry)(Reference Jacob, Gretz and Taylor2, Reference Pizzolo, Blom and Choi3). Another study in which the Illumina 450k array had been used(Reference Arpón, Riezu-Boj and Milagro4) was incorrectly not labelled as genome-wide. In addition, studies that used LINE-1 pyrosequencing, a common method for assessment of global methylation, were not labelled as global methylation studies (n 11) by the authors. These misclassifications highlight the difficulty in classifying studies when the type of assessment technique is not explicitly stated and the limitation of study heterogeneity as touched upon by ElGendy et al.(Reference ElGendy, Malcomson and Lara1).
To move the nutritional epigenetics field forward, we would like to propose a preferred choice for characterising dietary exposure and DNA methylation outcomes. In terms of dietary exposure, the field of nutritional epigenetics is currently dominated by studies evaluating supplements, despite the increased recognition that studying nutrients and foods in isolation neglects the complex combinations of nutrients that are likely synergistic and highly interactive(Reference Hu5). Disregarding nutrient interactions may be particularly naïve in relation to epigenetics as combinations of nutrients can increase or dilute the effects on DNA methylation. Therefore, we propose that dietary patterns should be the preferred dietary exposure to be evaluated. In terms of DNA methylation outcomes, the different DNA methylation assessment techniques available pose challenges to study design. Global methylation techniques are particularly suited when one expects large differences between the groups of interest, which may not be as relevant in the field of nutritional epigenetics. Candidate studies include genes selected based on a priori hypothesis regarding a trait of interest. This type of study is appealing because of low cost and straightforward statistical analyses(Reference Shabalin and Aberg6, Reference Kurdyukov and Bullock7), but it does not allow detection of novel loci. The use of genome-wide approaches in nutritional epigenetics is relatively new, with the first nutritional epigenome-wide association study (EWAS) included in the review published in 2010 using a 15k array and a sample size of 14(Reference Bouchard, Rabasa-Lhoret and Faraj8). The number of published EWAS has increased substantially since then with the launch of the 450k array in 2011, but application in the field of nutritional epigenetics has been limited to date (7/60 included studies). As genome-wide approaches allow for detection of novel loci, this type of DNA methylation assessment technique is particularly desirable when trying to understand the effects of diet on DNA methylation and should thus be recommended for the field of nutritional epigenetics, seeing this as a field in its infancy.
ElGendy et al. (Reference ElGendy, Malcomson and Lara1) concluded that there is little evidence for the effects of dietary factors other than folic acid on DNA methylation. They highlight the need for standardisation of DNA methylation assessment techniques. In addition to this recommendation, we would like to argue that there is a clear need for dietary randomised controlled trials using genome-wide DNA methylation assessment approaches with sufficiently large sample sizes, preferably evaluating interventions with dietary patterns rather than specific foods or nutrients in isolation. The time has come for concerted efforts and precise reporting in the nutritional epigenetics field.
Conflict of interest
None.




