The prevention of lifestyle-related diseases such as CVD, diabetes and stroke has attracted worldwide interest. Current treatment regimens for lifestyle-related diseases have shifted their focus onto the functionality of natural chemicals present in foods and drinks. One example is the association between the long-term consumption of red wine and reduced risk for CVD. In this context, resveratrol, a phytoalexin and antioxidant polyphenol present in red wine and various plants, has emerged as one of the most attractive and extensively studied compounds(Reference Lastra and Villegas1, Reference Baur and Sinclair2).
We have previously demonstrated that resveratrol suppresses the expression of cyclo-oxygenase (COX)-2, the rate-limiting enzyme in PG biosynthesis(Reference Smith3), particularly in 184B5/HER(184B5/human EGFR-related-2)-transformed mammary epithelial cells(Reference Subbaramaiah, Chung and Michaluart4). Additionally, we have shown that resveratrol activates PPARα, PPARβδ and PPARγ(Reference Inoue, Jiang and Katayama5, Reference Tsukamoto, Nakata and Tamura6), which are members of the nuclear receptor family of ligand-dependent transcription factors(Reference Michalik, Auwerx and Berger7, Reference Sonoda, Pei and Evans8); that it protects the brain against ischaemic stroke in mice through a PPARα-dependent mechanism demonstrated in cell-based reporter assays(Reference Inoue, Jiang and Katayama5) and that it acts via a negative feedback loop mediated through PPARγ to inhibit the expression of COX-2, particularly in macrophages(Reference Inoue, Tanabe and Umesono9). Based on these data, we have focused our attention on PPAR as potential molecular targets of resveratrol in the prevention of lifestyle-related diseases, the molecular mechanisms of which remain to be determined.
Previous studies have investigated the effects of resveratrol on the activation of silent mating type information regulation 2 homologue 1 (SIRT1), a NAD+-dependent protein deacetylase(Reference Howitz, Bitterman and Cohen10–Reference Lagouge, Argmann and Gerhart-Hines12), although a direct activation was not observed(Reference Kaeberlein, McDonagh and Heltweg13, Reference Pacholec, Bleasdale and Chrunyk14). Resveratrol has been reported to enhance the expression of endothelial NO synthase (eNOS)(Reference Wallerath, Deckert and Ternes15), which may be important for elucidating the effects of resveratrol on the cardiovascular system. However, these observations were based on human umbilical vein endothelial cells (HUVEC) treated with 33 μm-resveratrol for 24 h, conditions which are neither long term in nature nor physiologically relevant, and a time (24–72 h)- and concentration-dependent up-regulation of the expression of eNOS (2·8-fold in 100 μm-resveratrol) was observed not in HUVEC but in the EA.hy926 cell line, which is a hybrid of the HUVEC and human lung carcinoma A549 cells(Reference Wallerath, Deckert and Ternes15).
In the present study, we investigated the effects of resveratrol on the expression of eNOS using a physiological concentration (1 μm) during repeated treatments for 6 d. We demonstrated that the expression of eNOS and SIRT1 is up-regulated by resveratrol. We screened the differentially regulated genes in HUVEC on resveratrol treatment using microarray analysis and confirmed the candidate genes using quantitative RT-PCR analysis. We demonstrated that resveratrol increases the expression of autophagy-related genes, γ-aminobutyric acid A receptor-associated protein (GABARAP), microtubule-associated protein 1 light chain 3B (LC3B) and autophagy-related protein 3 (ATG3), and the radical scavenger activity-related metallothionein-1X (MT1X) gene. Additionally, we demonstrated that the expression of the cell-cycle checkpoint control RAD9 homologue B (RAD9B) gene is down-regulated by resveratrol. These data help in the identification of molecular mechanisms that may contribute to the beneficial effects of resveratrol on the cardiovascular system.
Experimental methods
Cell culture
HUVEC were obtained from the Japanese Collection of Research Bioresources and were grown in an endothelial cell growth medium (Cell Applications, Inc.). The cells were cultured for 6 d in the presence or absence of 1 μm-resveratrol or 1 μm-fenofibrate, a synthetic PPARα agonist, with the media being changed at 0, 2 and 4 d.
RNA extraction and analysis
Total RNA was isolated from the HUVEC using the acid guanidinium thiocyanate procedure and was analysed for gene expression via real-time quantitative RT-PCR (Mx3005; Stratagene) as described previously(Reference Hotta, Nakata and Katsukawa16). The primer pairs for the genes used in the present study and the cycling conditions are given in Table S1 (available online). The expression level of each mRNA was normalised to that of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA, which has been used for our previous study in HUVEC(Reference Inoue, Jiang and Katayama5, Reference Inoue, Taba and Miwa17)and bovine arterial endothelial cells (BAEC)(Reference Inoue, Tanabe and Umesono9, Reference Hotta, Nakata and Katsukawa16).
DNA microarray analysis
HUVEC were cultured for 6 d in the presence or absence of 1 μm-resveratrol. DNA microarray analysis was performed as described previously(Reference Shirotani-Ikejima, Kokame and Hamuro18). Briefly, poly(A)+ RNA was prepared from resveratrol-treated and untreated HUVEC using a QuickPrep micro mRNA purification kit (GE Healthcare) and was used to prepare complementary RNA samples as described in the GeneChip Expression Analysis Technical Manual (Affymetrix). Double-stranded complementary DNA was synthesised from 0·6 μg of poly(A)+ RNA using a MessageAMP™ II aRNA amplification kit (PE Applied Biosystems) with T7-(dT)24 primer (GE Healthcare). The product served as a template for the synthesis of biotin-labelled complementary RNA via in vitro transcription using a GeneChip IVT labelling kit (Affymetrix), and the amplified complementary RNA was fragmented by heat treatment in the presence of potassium acetate and magnesium acetate. The complementary RNA sample (10 μg) was then applied to a GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix), and hybridisation, washing, staining and scanning were performed according to the GeneChip Manual. The data for each probe set were calculated from the scanned array image using the GeneChip Analysis Suite (Affymetrix) and GeneSpring (Agilent Technologies). The procedure of analysis was as follows: for background subtraction, the median value of the negative control genes was subtracted from the raw values for each gene, and the resulting values were divided by the value of the 50th percentile of each chip to normalise the various signal intensities among the arrays. Finally, the ratios (resveratrol-treated:untreated and untreated:resveratrol-treated) were calculated from the average of duplicate normalised signal intensities. A change in expression of 1·5-fold was used as a threshold for comparison.
Statistical analysis
All results are expressed as the means and standard deviations. Comparisons between the groups were made using one-way ANOVA with post hoc Bonferroni multiple-comparison test or the unpaired t test. A P value < 0·05 was considered to indicate statistical significance.
Results and discussion
Repeated treatments with resveratrol up-regulate the expression of endothelial nitric oxide synthase
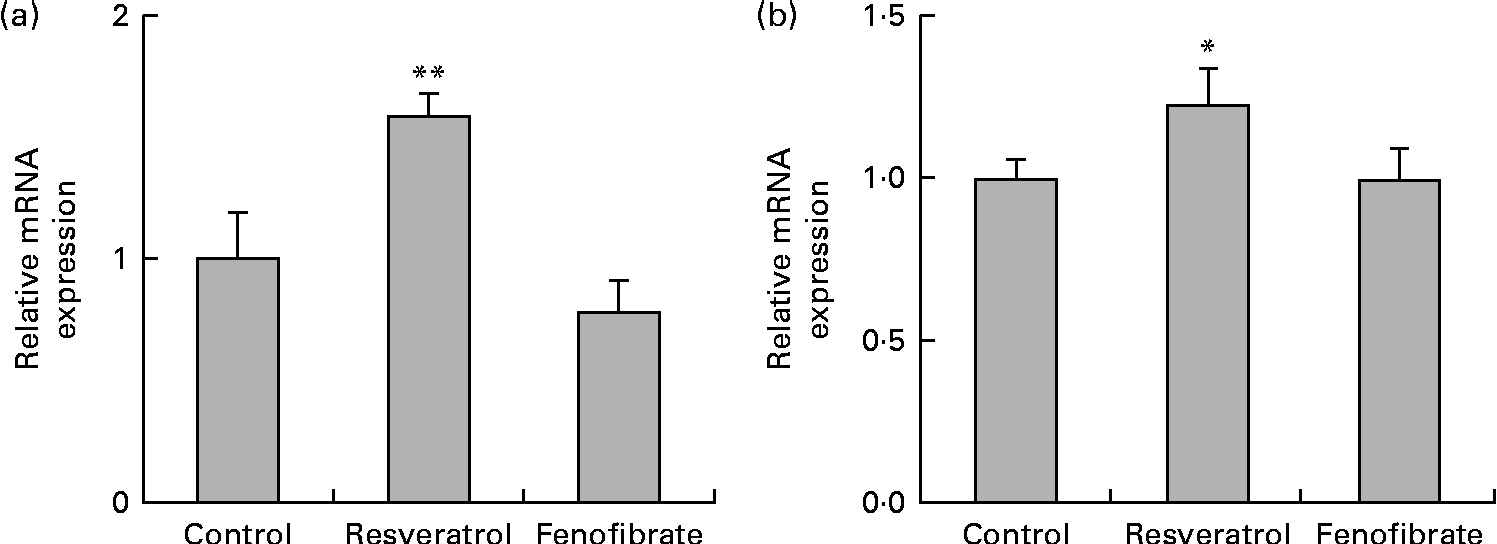
HUVEC were cultured for 6 d in the presence or absence of 1 μm-resveratrol and examined for the expression of eNOS by quantitative RT-PCR (Fig. 1(a)). The levels of eNOS mRNA were up-regulated 1·5-fold. Previous studies have demonstrated approximately 1·4-fold up-regulation of eNOS mRNA levels in HUVEC following treatment with 33 μm-resveratrol, but not with 1 μm, for 24 h(Reference Wallerath, Deckert and Ternes15), indicating that long-term repeated treatments with 1 μm-resveratrol for 6 d, as done in the present study, lead to the up-regulation of eNOS mRNA levels. Our results are in agreement with the report that daily treatment with resveratrol significantly increases the production of both eNOS protein and NO in HUVEC(Reference Takahashi and Nakashima19). Thus, induction of the expression of eNOS by repeated treatments with a physiological concentration of resveratrol can account, in part, for the cardiovascular benefits of the long-term consumption of red wine.

Fig. 1 Up-regulation of the expression of endothelial nitric oxide synthase (eNOS) and silent mating type information regulation 2 homologue 1 (SIRT1) mRNA and protein in human umbilical vein endothelial cells (HUVEC) following repeated treatments with 1 μm-resveratrol for 6 d. HUVEC were cultured for 6 d in the presence or absence of 1 μm-resveratrol or 1 μm-fenofibrate. Isolated RNA from the HUVEC was used for quantitative RT-PCR analysis. The mRNA levels of (a) eNOS and (b) SIRT1 were normalised to that of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. The normalised levels of eNOS and SIRT1 mRNA in the absence of resveratrol are expressed as 1, respectively. Values are means of three separate dishes, with standard deviations represented by vertical bars. Mean values were significantly different from those of the control and fenofibrate treatments: * P< 0·05 and ** P< 0·01 (one-way ANOVA with post hoc Bonferroni multiple-comparison test). Similar results were obtained by two additional experiments.
Repeated treatments with resveratrol up-regulate the expression of silent mating type information regulation 2 homologue 1
We examined the expression levels of SIRT1 mRNA (Fig. 1(b)) in HUVEC treated with 1 μm-resveratrol for 6 d. The levels of SIRT1 mRNA were up-regulated by 1·2-fold. As previous studies have reported that resveratrol is not a direct activator of SIRT1(Reference Kaeberlein, McDonagh and Heltweg13, Reference Pacholec, Bleasdale and Chrunyk14), we sought to investigate the mechanism of the up-regulation of SIRT1 levels. We have previously demonstrated that resveratrol is a triple agonist for PPARα, PPARβ/δ and PPARγ in vitro (Reference Inoue, Jiang and Katayama5, Reference Tsukamoto, Nakata and Tamura6) and that 4-week intake of resveratrol as well as the synthetic PPARα agonist fenofibrate up-regulates the hepatic expression of SIRT1 in wild-type, but not in PPARα knockout, mice (R Nakata et al., unpublished results). Therefore, we examined the expression of SIRT1 in HUVEC treated with 1 μm-fenofibrate for 6 d and observed that SIRT1 mRNA levels were unregulated by fenofibrate (Fig. 1(b)). Similarly, no effect on the expression of eNOS was observed following fenofibrate treatment (Fig. 1(a)). These discrepancies between the present study using HUVEC in vitro and the PPARα knockout mice study in vivo indicate the limitations of the present study. Cell culture of HUVEC is more difficult than that of BAEC since the former need several special supplements such as epidermal growth factor, basic fibroblast growth factor and vascular endothelial growth factor. Moreover, there are no interactions between HUVEC and other types of cells, which is common in vivo. On the other hand, fasting is reported to promote the expression of SIRT1 via the activation of PPARα in mice(Reference Hayashida, Arimoto and Kuramoto20), whereas there is no report of a connection between the up-regulation of eNOS levels and the activation of PPARα. Therefore, we could not answer why the expression of eNOS and SIRT1 was induced without involving PPAR due to the limitations of the present study.
Repeated treatments with resveratrol up-regulate the expression of autophagy-, radical scavenger activity- and anti-inflammatory activity-related genes and down-regulate the expression of a cell-cycle checkpoint control gene
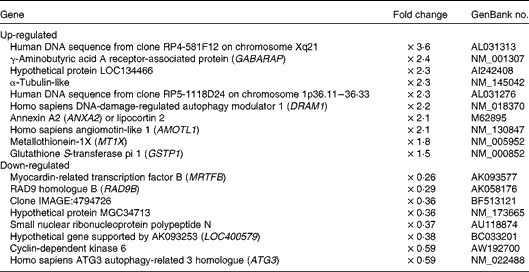
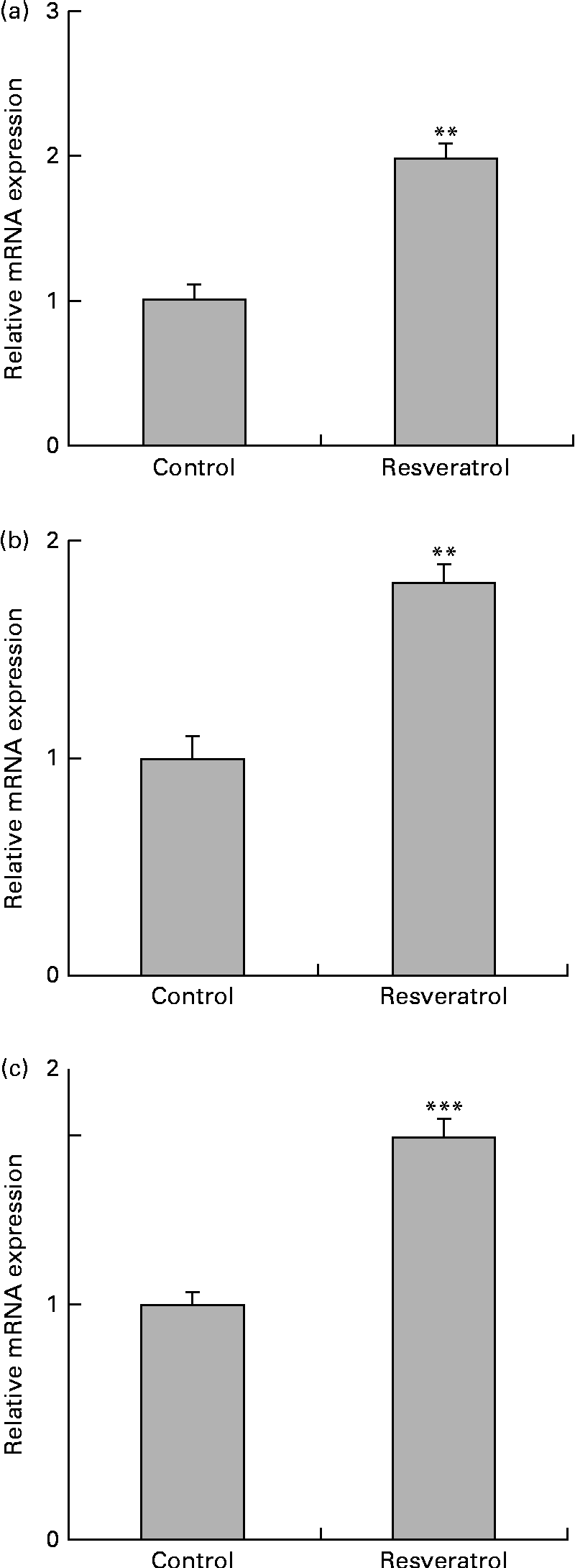
To examine the expression of other differentially regulated genes following resveratrol treatment, DNA microarray analysis was performed. Using cut-off values of 1·5-fold for induction and 0·67-fold for suppression, 256 up-regulated and 411 down-regulated genes were identified. Among these candidate genes (Table 1), the up-regulation of the autophagy-related GABARAP gene(Reference Mohrlüder, Schwarten and Willbold21) was confirmed by real-time quantitative RT-PCR (Fig. 2(a)). The induction of autophagocytosis by 50 μm-resveratrol and the extension of the autophagy-mediated lifespan have been reported previously(Reference Opipari, Tan and Boitano22, Reference Morselli, Galluzzi and Kepp23). In this context, we found that the expression of the autophagy-related LC3B and AGT3 genes was also up-regulated by treatment with 1 μm-resveratrol for 6 d (Fig. 2(b) and (c)), although these were not detected in the microarray analysis. Autophagy plays key roles in adaptation to stress such as starvation and normal development of the immune system as well as in a wide range of disease states. Therefore, the beneficial effects of resveratrol could partly be explained by the up-regulation of the expression of autophagy-related genes (Fig. 2). On the other hand, the beneficial effects of fenofibrate on retinal pigment epithelium have recently been reported to be partly due to the induction of autophagy(Reference Miranda, González-Rodríguez and García-Ramírez24); however, we could not observe the induction of the expression of autophagy-related genes by fenofibrate under our assay condition (data not shown).
Table 1 Representative genes differentially expressed following repeated treatments with resveratrol


Fig. 2 Up-regulation of the expression of autophagy-related genes in human umbilical vein endothelial cells (HUVEC) following repeated treatments with 1 μm-resveratrol for 6 d. HUVEC were cultured in the presence or absence of 1 μm-resveratrol as described in Fig. 1, and isolated mRNA from the HUVEC was used for quantitative RT-PCR analysis. The level of each mRNA was normalised to that of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. The normalised levels of the control mRNA are expressed as 1. The relative mRNA expression levels of (a) γ-aminobutyric acid A receptor-associated protein (GABARAP), (b) microtubule-associated protein 1 light chain 3B (LC3B) and (c) autophagy-related protein 3 (ATG3) are shown. Values are means of three separate dishes, with standard deviations represented by vertical bars. Mean values were significantly different from those of control: ** P< 0·01 and *** P< 0·001 (unpaired t test). Similar results were obtained by two additional experiments.
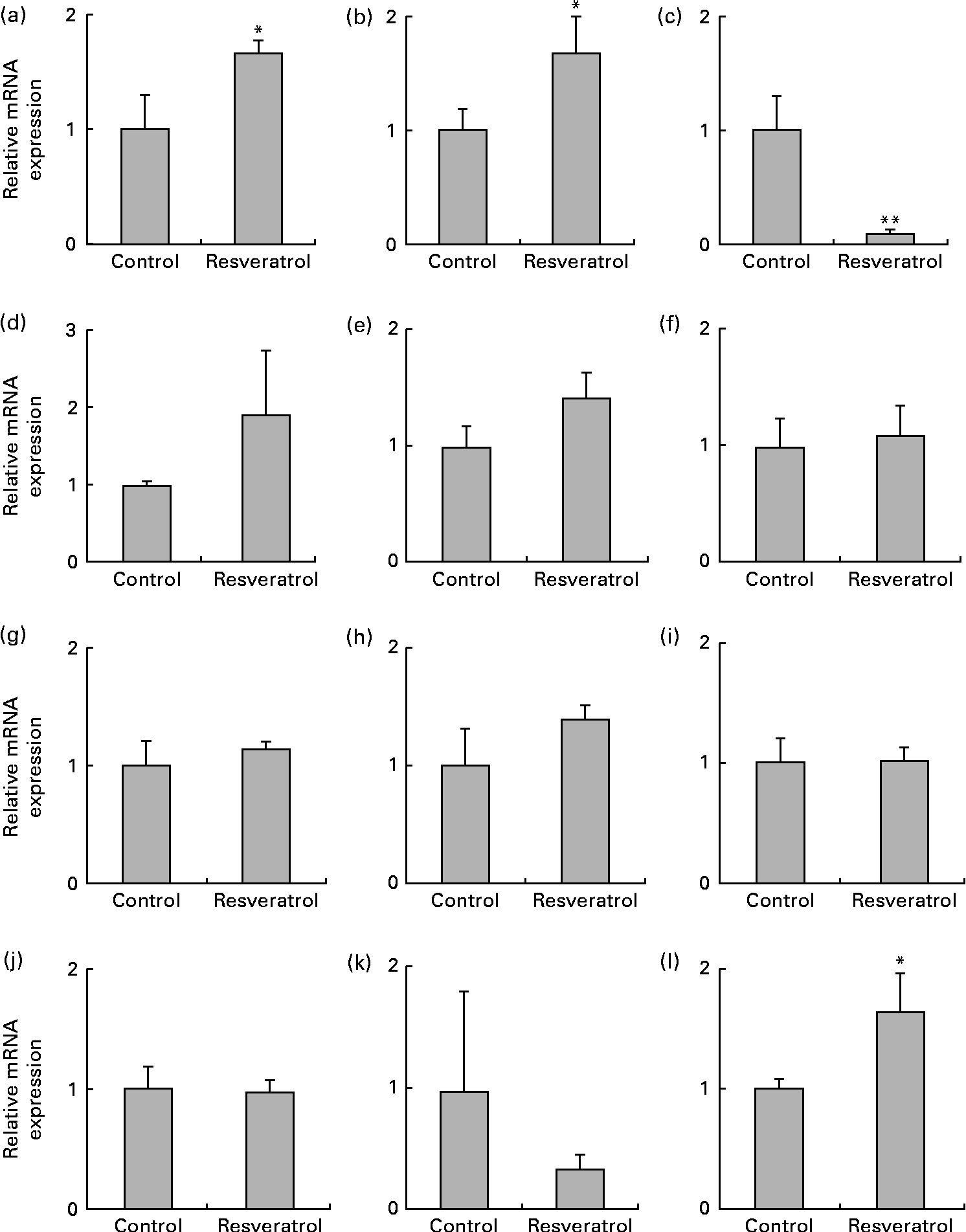
The up-regulation of the expression of the radical scavenger activity-related MT1X (Fig. 3(a)) and anti-inflammatory activity-related annexin A2 (ANXA2; Fig. 3(b)) genes and the down-regulation of the expression the RAD9B gene (Fig. 3(c)) were also observed using real-time quantitative RT-PCR analysis. ANXA2 is reported to be a regulator of cell surface plasmin generation(Reference Ling, Jacovina and Deora25), and there are no reports on the function of RAD9B, but its gene is a homologue of RAD9, an evolutionarily conserved gene with multiple functions for preserving genomic integrity(Reference Lieberman26), indicating that the beneficial effects of resveratrol may partly be due to the up-regulation of the expression of the antioxidant MT1X and anti-inflammatory ANXA2 genes and the down-regulation of the expression of the RAD9B gene. However, there were several discrepancies between the DNA microarray (Table 1) and real-time quantitative RT-PCR results for the angiomotin-like 1 (AMOTL1; Fig. 3(d)), LOC134466 (Fig. 3(e)), glutathione S-transferase pi 1 (GSTP1; Fig. 3(f)), myocardin-related transcription factor B (MRTFB; Fig. 3(g)) and cyclin-dependent kinase 6 (CDK6; Fig. 3(h)) genes. Similarly, induction of the expression of the eNOS and SIRT1 genes by resveratrol was detected using real-time quantitative RT-PCR (Fig. 1) but not by the microarray analysis. These results show that the DNA microarray analysis is useful for the screening of candidate genes, but this should be confirmed by real-time quantitative RT-PCR.

Fig. 3 Expression of the radical scavenger activity-, anti-inflammatory activity- and cell-cycle checkpoint control-related genes and cyclo-oxygenase 2 (COX-2), PPARα, PPARβ/δ and PPARγ genes in human umbilical vein endothelial cells (HUVEC) following repeated treatments with 1 μm-resveratrol for 6 d. The mRNA was isolated from the HUVEC for quantitative RT-PCR analysis following treatment with 1 μm-resveratrol for 6 d. The level of each mRNA was normalised to that of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. The relative mRNA expression levels of (a) metallothionein-1X (MT1X), (b) anti-inflammatory activity-related annexin A2 (ANXA2), (c) RAD9 homologue B (RAD9B), (d) angiomotin-like 1 (AMOTL1), (e) LOC134466, (f) glutathione S-transferase pi 1 (GSTP1), (g) myocardin-related transcription factor B (MRTFB), (h) cyclin-dependent kinase 6 (CDK6), (i) COX-2, (j) PPARα, (k) PPARβ/δ and (l) PPARγ are shown. Values are means of three separate dishes, with standard deviations represented by vertical bars. Mean values were significantly different from those of control: * P< 0·05 and ** P< 0·01 (unpaired t test). Similar results were obtained by two additional experiments.
Finally, we examined the expression of COX-2 and PPAR. The expression of COX-2 was unchanged following resveratrol treatment (Fig. 3(i)). This is consistent with our previous observations that resveratrol suppresses the expression of COX-2 in 184B5/HER-transformed mammary epithelial cells(Reference Subbaramaiah, Chung and Michaluart4), but not in BAEC (H Inoue, unpublished results), and that 15-deoxy-Δ12,14 PGJ2, a natural PPARγ agonist, suppresses the expression of COX-2 in macrophage-like U937 cells, but not in BAEC(Reference Inoue, Tanabe and Umesono9). The expression levels of PPARα (Fig. 3(j)) and PPARβ/δ (Fig. 3(k)) were also unchanged following resveratrol treatment; however, the expression of PPARγ was up-regulated (Fig. 3(l)). Increased expression of PPARγ had not been detected previously in BAEC(Reference Inoue, Tanabe and Umesono9), indicating possible differences between these cell lines.
In summary, we demonstrated that the expression of eNOS and SIRT1 is up-regulated in HUVEC following repeated treatments with a physiological concentration (1 μm) of resveratrol for 6 d. These results will account, in part, for the cardiovascular benefits of the long-term consumption of red wine. Moreover, genes related to autophagy, radical scavenger activity, anti-inflammatory activity and cell-cycle checkpoint control were shown to be differentially regulated in this condition. Modulating the expression of these genes may contribute to the observed beneficial effects of resveratrol on the cardiovascular system.
Supplementary material
To view supplementary material for this article, please visit http://dx.doi.org/10.1017/S0007114513001670
Acknowledgements
We thank Dr Hiroko Shirotani-Ikejima, Dr Michiko Katsukawa, Ms Kumiko Satoh, Ms Chisako Ogura and Ms Naoko Anzai for their technical assistance. The present study was supported by a Grant-in-Aid for Scientific Research (no. 19300250 and 24300217 to H. I. and R. N.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, the Iijima Memorial Foundation for the Promotion of Food Science and Technology, and Japan Food Chemical Research Foundation. H. I., T. Y., R. Y. and R. N. designed the research. Y. T., Y. K., H. A., E. T. and A. T. performed the experiments and analysed the data. K. K. and T. M. coordinated the DNA microarray analysis. H. I., Y. T., Y. K. and R. N. discussed the results and wrote the manuscript. The authors declare that there are no conflicts of interest.